OXA β-lactamases
- PMID: 24696435
- PMCID: PMC3993105
- DOI: 10.1128/CMR.00117-13
OXA β-lactamases
Abstract
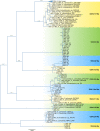
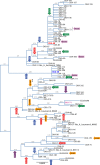
The OXA β-lactamases were among the earliest β-lactamases detected; however, these molecular class D β-lactamases were originally relatively rare and always plasmid mediated. They had a substrate profile limited to the penicillins, but some became able to confer resistance to cephalosporins. From the 1980s onwards, isolates of Acinetobacter baumannii that were resistant to the carbapenems emerged, manifested by plasmid-encoded β-lactamases (OXA-23, OXA-40, and OXA-58) categorized as OXA enzymes because of their sequence similarity to earlier OXA β-lactamases. It was soon found that every A. baumannii strain possessed a chromosomally encoded OXA β-lactamase (OXA-51-like), some of which could confer resistance to carbapenems when the genetic environment around the gene promoted its expression. Similarly, Acinetobacter species closely related to A. baumannii also possessed their own chromosomally encoded OXA β-lactamases; some could be transferred to A. baumannii, and they formed the basis of transferable carbapenem resistance in this species. In some cases, the carbapenem-resistant OXA β-lactamases (OXA-48) have migrated into the Enterobacteriaceae and are becoming a significant cause of carbapenem resistance. The emergence of OXA enzymes that can confer resistance to carbapenems, particularly in A. baumannii, has transformed these β-lactamases from a minor hindrance into a major problem set to demote the clinical efficacy of the carbapenems.
Figures






References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

