Reduced synaptic STIM2 expression and impaired store-operated calcium entry cause destabilization of mature spines in mutant presenilin mice
- PMID: 24698269
- PMCID: PMC4007018
- DOI: 10.1016/j.neuron.2014.02.019
Reduced synaptic STIM2 expression and impaired store-operated calcium entry cause destabilization of mature spines in mutant presenilin mice
Abstract
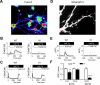
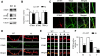
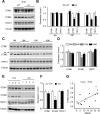
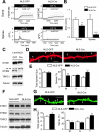
Mushroom dendritic spine structures are essential for memory storage, and the loss of mushroom spines may explain memory defects in Alzheimer's disease (AD). Here we show a significant reduction in the fraction of mushroom spines in hippocampal neurons from the presenilin-1 M146V knockin (KI) mouse model of familial AD (FAD). The stabilization of mushroom spines depends on STIM2-mediated neuronal store-operated calcium influx (nSOC) and continuous activity of Ca(2+)/calmodulin-dependent protein kinase II (CaMKII). We demonstrate that STIM2-nSOC-CaMKII pathway is compromised in KI neurons, in aging neurons, and in sporadic AD brains due to downregulation of STIM2 protein. We further establish that overexpression of STIM2 rescues synaptic nSOC, CaMKII activity, and mushroom spine loss in KI neurons. Our results identify STIM2-nSOC-CaMKII synaptic maintenance pathway as a novel potential therapeutic target for treatment of AD and age-related memory decline.
Copyright © 2014 Elsevier Inc. All rights reserved.
Figures



Similar articles
-
Dysregulation of neuronal calcium homeostasis in Alzheimer's disease - A therapeutic opportunity?Biochem Biophys Res Commun. 2017 Feb 19;483(4):998-1004. doi: 10.1016/j.bbrc.2016.09.053. Epub 2016 Sep 15. Biochem Biophys Res Commun. 2017. PMID: 27641664 Free PMC article. Review.
-
Neuronal Store-Operated Calcium Entry and Mushroom Spine Loss in Amyloid Precursor Protein Knock-In Mouse Model of Alzheimer's Disease.J Neurosci. 2015 Sep 30;35(39):13275-86. doi: 10.1523/JNEUROSCI.1034-15.2015. J Neurosci. 2015. PMID: 26424877 Free PMC article.
-
Store-Operated Calcium Channel Complex in Postsynaptic Spines: A New Therapeutic Target for Alzheimer's Disease Treatment.J Neurosci. 2016 Nov 23;36(47):11837-11850. doi: 10.1523/JNEUROSCI.1188-16.2016. J Neurosci. 2016. PMID: 27881772 Free PMC article.
-
Stim2-Eb3 Association and Morphology of Dendritic Spines in Hippocampal Neurons.Sci Rep. 2017 Dec 15;7(1):17625. doi: 10.1038/s41598-017-17762-8. Sci Rep. 2017. PMID: 29247211 Free PMC article.
-
Exploring the Role of PSEN Mutations in the Pathogenesis of Alzheimer's Disease.Neurotox Res. 2020 Dec;38(4):833-849. doi: 10.1007/s12640-020-00232-x. Epub 2020 Jun 18. Neurotox Res. 2020. PMID: 32556937 Review.
Cited by
-
Dysregulation of neuronal calcium homeostasis in Alzheimer's disease - A therapeutic opportunity?Biochem Biophys Res Commun. 2017 Feb 19;483(4):998-1004. doi: 10.1016/j.bbrc.2016.09.053. Epub 2016 Sep 15. Biochem Biophys Res Commun. 2017. PMID: 27641664 Free PMC article. Review.
-
The sigma-1 receptor mediates the beneficial effects of pridopidine in a mouse model of Huntington disease.Neurobiol Dis. 2017 Jan;97(Pt A):46-59. doi: 10.1016/j.nbd.2016.10.006. Epub 2016 Nov 3. Neurobiol Dis. 2017. PMID: 27818324 Free PMC article.
-
Identification of molecular determinants that govern distinct STIM2 activation dynamics.PLoS Biol. 2018 Nov 16;16(11):e2006898. doi: 10.1371/journal.pbio.2006898. eCollection 2018 Nov. PLoS Biol. 2018. PMID: 30444880 Free PMC article.
-
Role of STIM1 in neurodegeneration.World J Biol Chem. 2018 Nov 16;9(2):16-24. doi: 10.4331/wjbc.v9.i2.16. World J Biol Chem. 2018. PMID: 30568747 Free PMC article. Review.
-
STIM Protein-NMDA2 Receptor Interaction Decreases NMDA-Dependent Calcium Levels in Cortical Neurons.Cells. 2020 Jan 9;9(1):160. doi: 10.3390/cells9010160. Cells. 2020. PMID: 31936514 Free PMC article.
References
-
- Akbari Y, Hitt BD, Murphy MP, Dagher NN, Tseng BP, Green KN, Golde TE, LaFerla FM. Presenilin regulates capacitative calcium entry dependently and independently of gamma-secretase activity. Biochemical and biophysical research communications. 2004;322:1145–1152. - PubMed
-
- Berna-Erro A, Braun A, Kraft R, Kleinschnitz C, Schuhmann MK, Stegner D, Wultsch T, Eilers J, Meuth SG, Stoll G, et al. STIM2 regulates capacitive Ca2+ entry in neurons and plays a key role in hypoxic neuronal cell death. Sci Signal. 2009;2:ra67. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

