Multi-marker analysis of circulating cell-free DNA toward personalized medicine for colorectal cancer
- PMID: 24698732
- PMCID: PMC5528519
- DOI: 10.1016/j.molonc.2014.02.005
Multi-marker analysis of circulating cell-free DNA toward personalized medicine for colorectal cancer
Abstract
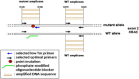
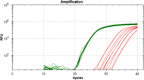
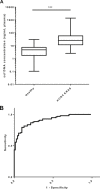
Development of a Q-PCR-based assay for the high-performance analysis of circulating cell-free DNA (ccfDNA) requires good knowledge of its structure and size. In this work, we present the first visual determination of ccfDNA by Atomic Force Microscopy (AFM) on plasma samples from colorectal cancer (CRC) patients and healthy donors. In addition to the examination of fragment size distribution profile as performed by Q-PCR, this analysis confirms that ccfDNA is highly fragmented and that more than 80% of ccfDNA fragments in CRC plasma are below 145 bp. We adapted an Allele-Specific Blocker (ASB) Q-PCR to small ccfDNA fragments to determine simultaneously the total ccfDNA concentration, the presence of point mutation, the proportion of mutated allele, and a ccfDNA integrity index. The data validated analytically these four parameters in 124 CRC clinical samples and 71 healthy individuals. The multi-marker method, termed Intplex, enables sensitive and specific non-invasive analysis of tumor ccfDNA, which has great potential in terms of cost, quality control, and easy implementation in every clinical center laboratory.
Keywords: Circulating cell-free DNA; Colorectal cancer; Multi-marker analysis; qPCR.
Copyright © 2014 Federation of European Biochemical Societies. Published by Elsevier B.V. All rights reserved.
Figures



References
-
- Amado, R.G. , Wolf, M. , Peeters, M. , Van Cutsem, E. , Siena, S. , Freeman, D.J. , Juan, T. , Sikorski, R. , Suggs, S. , Radinsky, R. , Patterson, S.D. , Chang, D.D. , 2008. Wild-type KRAS is required for panitumumab efficacy in patients with metastatic colorectal cancer. J. Clin. Oncol.. 26, (10) 1626–1634. - DOI - PubMed
-
- Bamford, S. , Dawson, E. , Forbes, S. , Clements, J. , Pettett, R. , Dogan, A. , Flanagan, A. , Teague, J. , Futreal, P.A. , Stratton, M.R. , Wooster, R. , 2004. The COSMIC (catalogue of somatic mutations in cancer) database and website. Br. J. Cancer. 91, (2) 355–358. DOI: 6601894, [pii] 10.1038/sj.bjc.6601894 - PMC - PubMed
-
- Bustin, S.A. , Benes, V. , Garson, J.A. , Hellemans, J. , Huggett, J. , Kubista, M. , Mueller, R. , Nolan, T. , Pfaffl, M.W. , Shipley, G.L. , Vandesompele, J. , Wittwer, C.T. , 2009. The MIQE guidelines: minimum information for publication of quantitative real-time PCR experiments. Clin. Chem.. 55, (4) 611–622. [pii] 10.1373/clinchem.2008.112797, DOI: clinchem.2008.112797 - PubMed
-
- Chan, K.C. , Jiang, P. , Zheng, Y.W. , Liao, G.J. , Sun, H. , Wong, J. , Siu, S.S. , Chan, W.C. , Chan, S.L. , Chan, A.T. , Lai, P.B. , Chiu, R.W. , Lo, Y.M. , 2013. Cancer genome scanning in plasma: detection of tumor-associated copy number aberrations, single-nucleotide variants, and tumoral heterogeneity by massively parallel sequencing. Clin. Chem.. 59, (1) 211–224. - DOI - PubMed
-
- Dawson, S.J. , Tsui, D.W. , Murtaza, M. , Biggs, H. , Rueda, O.M. , Chin, S.F. , Dunning, M.J. , Gale, D. , Forshew, T. , Mahler-Araujo, B. , Rajan, S. , Humphray, S. , Becq, J. , Halsall, D. , Wallis, M. , Bentley, D. , Caldas, C. , Rosenfeld, N. , 2013. Analysis of circulating tumor DNA to monitor metastatic breast cancer. N. Engl. J. Med.. 368, (13) 1199–1209. - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

