Genomic evolution of porcine reproductive and respiratory syndrome virus (PRRSV) isolates revealed by deep sequencing
- PMID: 24698958
- PMCID: PMC3974674
- DOI: 10.1371/journal.pone.0088807
Genomic evolution of porcine reproductive and respiratory syndrome virus (PRRSV) isolates revealed by deep sequencing
Abstract
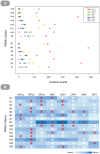
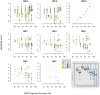
Most studies on PRRSV evolution have been limited to a particular region of the viral genome. A thorough genome-wide understanding of the impact of different mechanisms on shaping PRRSV genetic diversity is still lacking. To this end, deep sequencing was used to obtain genomic sequences of a diverse set of 16 isolates from a region of Hong Kong with a complex PRRSV epidemiological record. Genome assemblies and phylogenetic typing indicated the co-circulation of strains of both genotypes (type 1 and type 2) with varying Nsp2 deletion patterns and distinct evolutionary lineages ("High Fever"-like and local endemic type). Recombination analyses revealed genomic breakpoints in structural and non-structural regions of genomes of both genotypes with evidence of many recombination events originating from common ancestors. Additionally, the high fold of coverage per nucleotide allowed the characterization of minor variants arising from the quasispecies of each strain. Overall, 0.56-2.83% of sites were found to be polymorphic with respect to cognate consensus genomes. The distribution of minor variants across each genome was not uniform indicating the influence of selective forces. Proportion of variants capable of causing an amino acid change in their respective codons ranged between 25-67% with many predicted to be non-deleterious. Low frequency deletion variants were also detected providing one possible mechanism for their sudden emergence as cited in previous reports.
Conflict of interest statement
Figures


References
-
- Nodelijk G (2002) Porcine reproductive and respiratory syndrome (PRRS) with special reference to clinical aspects and diagnosis: a review. Veterinary quarterly 24: 96–100. - PubMed
-
- Hill H (1990) Overview and history of mystery swine disease (swine infertility/respiratory syndrome). PrOf Mystery Swine Dis Committee 29–31.
-
- Paton D (1995) Porcine reproductive and respiratory syndrome (blue-eared pig disease). Reviews in Medical Microbiology 6: 119.
-
- Edwards S, Robertson I, Wilesmith J, Ryan J, Kilner C, et al. (1992) PRRS (“blue-eared pig disease”) in Great Britain. AASP Newsletter 4: 32–36.
-
- Plana J, Vayreda M, Vilarrasa J, Bastons M, Rosell R, et al. (1992) Porcine epidemic abortion and respiratory syndrome (mystery swine disease). Isolation in Spain of the causative agent and experimental reproduction of the disease. Veterinary microbiology 33: 203–211. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

