IFITM3 restricts influenza A virus entry by blocking the formation of fusion pores following virus-endosome hemifusion
- PMID: 24699674
- PMCID: PMC3974867
- DOI: 10.1371/journal.ppat.1004048
IFITM3 restricts influenza A virus entry by blocking the formation of fusion pores following virus-endosome hemifusion
Abstract
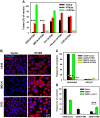
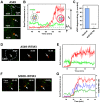
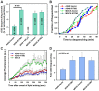
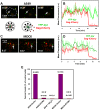
Interferon-induced transmembrane proteins (IFITMs) inhibit infection of diverse enveloped viruses, including the influenza A virus (IAV) which is thought to enter from late endosomes. Recent evidence suggests that IFITMs block virus hemifusion (lipid mixing in the absence of viral content release) by altering the properties of cell membranes. Consistent with this mechanism, excess cholesterol in late endosomes of IFITM-expressing cells has been reported to inhibit IAV entry. Here, we examined IAV restriction by IFITM3 protein using direct virus-cell fusion assay and single virus imaging in live cells. IFITM3 over-expression did not inhibit lipid mixing, but abrogated the release of viral content into the cytoplasm. Although late endosomes of IFITM3-expressing cells accumulated cholesterol, other interventions leading to aberrantly high levels of this lipid did not inhibit virus fusion. These results imply that excess cholesterol in late endosomes is not the mechanism by which IFITM3 inhibits the transition from hemifusion to full fusion. The IFITM3's ability to block fusion pore formation at a post-hemifusion stage shows that this protein stabilizes the cytoplasmic leaflet of endosomal membranes without adversely affecting the lumenal leaflet. We propose that IFITM3 interferes with pore formation either directly, through partitioning into the cytoplasmic leaflet of a hemifusion intermediate, or indirectly, by modulating the lipid/protein composition of this leaflet. Alternatively, IFITM3 may redirect IAV fusion to a non-productive pathway, perhaps by promoting fusion with intralumenal vesicles within multivesicular bodies/late endosomes.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures
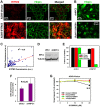
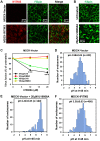
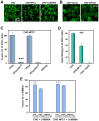
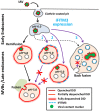
References
-
- Perreira JM, Chin CR, Feeley EM, Brass AL (2013) IFITMs Restrict the Replication of Multiple Pathogenic Viruses. J Mol Biol 425: 4937–55 doi:10.1016/j.jmb.2013.09.024 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

