Strain-specific nuclear genetic background differentially affects mitochondria-related phenotypes in Saccharomyces cerevisiae
- PMID: 24700775
- PMCID: PMC4082703
- DOI: 10.1002/mbo3.167
Strain-specific nuclear genetic background differentially affects mitochondria-related phenotypes in Saccharomyces cerevisiae
Abstract
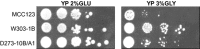
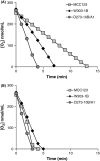
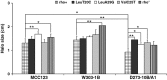
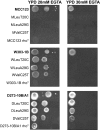
In the course of our studies on mitochondrial defects, we have observed important phenotypic variations in Saccharomyces cerevisiae strains suggesting that a better characterization of the genetic variability will be essential to define the relationship between the mitochondrial efficiency and the presence of different nuclear backgrounds. In this manuscript, we have extended the study of such relations by comparing phenotypic assays related to mitochondrial functions of three wild-type laboratory strains. In addition to the phenotypic variability among the wild-type strains, important differences have been observed among strains bearing identical mitochondrial tRNA mutations that could be related only to the different nuclear background of the cells. Results showed that strains exhibited an intrinsic variability in the severity of the effects of the mitochondrial mutations and that specific strains might be used preferentially to evaluate the phenotypic effect of mitochondrial mutations on carbon metabolism, stress responses, and mitochondrial DNA stability. In particular, while W303-1B and MCC123 strains should be used to study the effect of severe mitochondrial tRNA mutations, D273-10B/A1 strain is rather suitable for studying the effects of milder mutations.
Keywords: Diseases; nuclear background; oxidative stress; respiration; yeast model.
© 2014 The Authors. MicrobiologyOpen published by John Wiley & Sons Ltd.
Figures



Similar articles
-
Mitochondrial diseases: Yeast as a model for the study of suppressors.Biochim Biophys Acta Mol Cell Res. 2017 Apr;1864(4):666-673. doi: 10.1016/j.bbamcr.2017.01.008. Epub 2017 Jan 12. Biochim Biophys Acta Mol Cell Res. 2017. PMID: 28089773
-
Analyzing the suppression of respiratory defects in the yeast model of human mitochondrial tRNA diseases.Gene. 2013 Sep 15;527(1):1-9. doi: 10.1016/j.gene.2013.05.042. Epub 2013 May 30. Gene. 2013. PMID: 23727608
-
In Vivo Analysis of Mitochondrial Protein Synthesis in Saccharomyces cerevisiae Mitochondrial tRNA Mutants.Methods Mol Biol. 2022;2497:243-254. doi: 10.1007/978-1-0716-2309-1_15. Methods Mol Biol. 2022. PMID: 35771446
-
Mitochondria-nucleus network for genome stability.Free Radic Biol Med. 2015 May;82:73-104. doi: 10.1016/j.freeradbiomed.2015.01.013. Epub 2015 Jan 30. Free Radic Biol Med. 2015. PMID: 25640729 Review.
-
Mitochondrial Aminoacyl-tRNA Synthetase and Disease: The Yeast Contribution for Functional Analysis of Novel Variants.Int J Mol Sci. 2021 Apr 26;22(9):4524. doi: 10.3390/ijms22094524. Int J Mol Sci. 2021. PMID: 33926074 Free PMC article. Review.
Cited by
-
Gcn5 histone acetyltransferase is present in the mitoplasts.Biol Open. 2019 Feb 18;8(2):bio041244. doi: 10.1242/bio.041244. Biol Open. 2019. PMID: 30777878 Free PMC article.
-
Specificity Determination in Saccharomyces cerevisiae Killer Virus Systems.Microorganisms. 2021 Jan 23;9(2):236. doi: 10.3390/microorganisms9020236. Microorganisms. 2021. PMID: 33498746 Free PMC article.
-
Biolistic Approach for Transient Gene Expression Studies in Plants.Methods Mol Biol. 2020;2124:125-139. doi: 10.1007/978-1-0716-0356-7_6. Methods Mol Biol. 2020. PMID: 32277451 Free PMC article. Review.
References
-
- Baruffini E, Ferrero I, Foury F. Mitochondrial DNA defects in Saccharomyces cerevisiae caused by functional interactions between DNA polymerase gamma mutations associated with diseases in human. Biochim. Biophys. Acta. 2007;1772:1225–1235. - PubMed
-
- Carelli V, Giordano C, d'Amati G. Pathogenic expression of homoplasmic mtDNA mutations needs a complex nuclear-mitochondrial interaction. Trends Genet. 2003;19:257–262. - PubMed
-
- De Luca C, Besagni C, Frontali L, Bolotin-Fukuhara M, Francisci S. Mutations in yeast mt tRNAs: specific and general suppression by nuclear encoded tRNA interactors. Gene. 2006;377:169–176. - PubMed
-
- De Luca C, Zhou Y, Montanari A, Morea V, Oliva R, Besagni C, et al. Can yeast be used to study mitochondrial diseases? Biolistic tRNA mutants for the analysis of mechanisms and suppressors. Mitochondrion. 2009;9:408–417. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

