Comparative meta-analysis of prognostic gene signatures for late-stage ovarian cancer
- PMID: 24700801
- PMCID: PMC4580554
- DOI: 10.1093/jnci/dju049
Comparative meta-analysis of prognostic gene signatures for late-stage ovarian cancer
Abstract
Background: Ovarian cancer is the fifth most common cause of cancer deaths in women in the United States. Numerous gene signatures of patient prognosis have been proposed, but diverse data and methods make these difficult to compare or use in a clinically meaningful way. We sought to identify successful published prognostic gene signatures through systematic validation using public data.
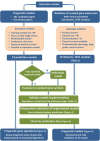
Methods: A systematic review identified 14 prognostic models for late-stage ovarian cancer. For each, we evaluated its 1) reimplementation as described by the original study, 2) performance for prognosis of overall survival in independent data, and 3) performance compared with random gene signatures. We compared and ranked models by validation in 10 published datasets comprising 1251 primarily high-grade, late-stage serous ovarian cancer patients. All tests of statistical significance were two-sided.
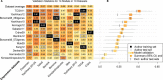
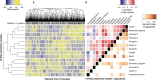
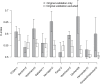
Results: Twelve published models had 95% confidence intervals of the C-index that did not include the null value of 0.5; eight outperformed 97.5% of signatures including the same number of randomly selected genes and trained on the same data. The four top-ranked models achieved overall validation C-indices of 0.56 to 0.60 and shared anticorrelation with expression of immune response pathways. Most models demonstrated lower accuracy in new datasets than in validation sets presented in their publication.
Conclusions: This analysis provides definitive support for a handful of prognostic models but also confirms that these require improvement to be of clinical value. This work addresses outstanding controversies in the ovarian cancer literature and provides a reproducible framework for meta-analytic evaluation of gene signatures.
© The Author 2014. Published by Oxford University Press. All rights reserved. For Permissions, please e-mail: journals.permissions@oup.com.
Figures

Comment in
-
Individualizing care for ovarian cancer patients using big data.J Natl Cancer Inst. 2014 Apr 3;106(5):dju080. doi: 10.1093/jnci/dju080. J Natl Cancer Inst. 2014. PMID: 24700802 No abstract available.
References
-
- Siegel R, Naishadham D, Jemal A. Cancer statistics, 2012. CA Cancer J Clin. 2012;62(1):10–29. - PubMed
-
- Cannistra SA, Bast RC, Jr, Berek JS, et al. Progress in the management of gynecologic cancer: consensus summary statement. J Clin Oncol. 2003;21(10 Suppl):129s–132s. - PubMed
-
- Na YJ, Farley J, Zeh A, et al. Ovarian cancer: markers of response. Int J Gynecol Cancer. 2009;19(Suppl 2):S21–S29. - PubMed
-
- Chon HS, Lancaster JM. Microarray-based gene expression studies in ovarian cancer. Cancer Control. 2011;18(1):8–15. - PubMed
-
- Sabatier R, Finetti P, Cervera N, et al. Gene expression profiling and prediction of clinical outcome in ovarian cancer. Crit Rev Oncol Hematol. 2009;72(2):98–109. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

