Pathways disrupted in human ALS motor neurons identified through genetic correction of mutant SOD1
- PMID: 24704492
- PMCID: PMC4653065
- DOI: 10.1016/j.stem.2014.03.004
Pathways disrupted in human ALS motor neurons identified through genetic correction of mutant SOD1
Abstract
Although many distinct mutations in a variety of genes are known to cause Amyotrophic Lateral Sclerosis (ALS), it remains poorly understood how they selectively impact motor neuron biology and whether they converge on common pathways to cause neuronal degeneration. Here, we have combined reprogramming and stem cell differentiation approaches with genome engineering and RNA sequencing to define the transcriptional and functional changes that are induced in human motor neurons by mutant SOD1. Mutant SOD1 protein induced a transcriptional signature indicative of increased oxidative stress, reduced mitochondrial function, altered subcellular transport, and activation of the ER stress and unfolded protein response pathways. Functional studies demonstrated that these pathways were perturbed in a manner dependent on the SOD1 mutation. Finally, interrogation of stem-cell-derived motor neurons produced from ALS patients harboring a repeat expansion in C9orf72 indicates that at least a subset of these changes are more broadly conserved in ALS.
Copyright © 2014 Elsevier Inc. All rights reserved.
Figures

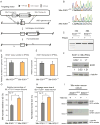
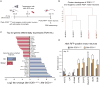




Comment in
-
The ER under rapid fire.EMBO J. 2014 Jun 2;33(11):1195-7. doi: 10.1002/embj.201488692. Epub 2014 Apr 30. EMBO J. 2014. PMID: 24788411 Free PMC article.
References
-
- Bilican B, Serio A, Barmada SJ, Nishimura AL, Sullivan GJ, Carrasco M, Phatnani HP, Puddifoot CA, Story D, Fletcher J, et al. Mutant induced pluripotent stem cell lines recapitulate aspects of TDP-43 proteinopathies and reveal cell-specific vulnerability. Proc Natl Acad Sci U S A. 2012;109:5803–5808. - PMC - PubMed
-
- Boyce M, Bryant KF, Jousse C, Long K, Harding HP, Scheuner D, Kaufman RJ, Ma D, Coen DM, Ron D, et al. A selective inhibitor of eIF2alpha dephosphorylation protects cells from ER stress. Science. 2005;307:935–939. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
- DP1 GM105378/GM/NIGMS NIH HHS/United States
- RC2 NS069395/NS/NINDS NIH HHS/United States
- 1U24NS078736-01/NS/NINDS NIH HHS/United States
- RC2 NS070342/NS/NINDS NIH HHS/United States
- HHMI/Howard Hughes Medical Institute/United States
- OD006862/OD/NIH HHS/United States
- KL2 TR000168/TR/NCATS NIH HHS/United States
- DP1 OD006862/OD/NIH HHS/United States
- R01 NS050557/NS/NINDS NIH HHS/United States
- 5RC2NS069395-02/NS/NINDS NIH HHS/United States
- RC2-NS070-342/NS/NINDS NIH HHS/United States
- 1R01NS050557/NS/NINDS NIH HHS/United States
- U24 NS078736/NS/NINDS NIH HHS/United States
- R00 NS077435/NS/NINDS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

