Modeling ALS with iPSCs reveals that mutant SOD1 misregulates neurofilament balance in motor neurons
- PMID: 24704493
- PMCID: PMC4230530
- DOI: 10.1016/j.stem.2014.02.004
Modeling ALS with iPSCs reveals that mutant SOD1 misregulates neurofilament balance in motor neurons
Abstract
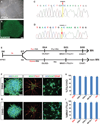
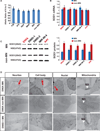
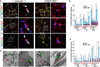
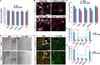
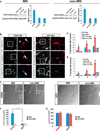
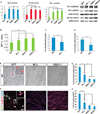
Amyotrophic lateral sclerosis (ALS) presents motoneuron (MN)-selective protein inclusions and axonal degeneration but the underlying mechanisms of such are unknown. Using induced pluripotent cells (iPSCs) from patients with mutation in the Cu/Zn superoxide dismutase (SOD1) gene, we show that spinal MNs, but rarely non-MNs, exhibited neurofilament (NF) aggregation followed by neurite degeneration when glia were not present. These changes were associated with decreased stability of NF-L mRNA and binding of its 3' UTR by mutant SOD1 and thus altered protein proportion of NF subunits. Such MN-selective changes were mimicked by expression of a single copy of the mutant SOD1 in human embryonic stem cells and were prevented by genetic correction of the SOD1 mutation in patient's iPSCs. Importantly, conditional expression of NF-L in the SOD1 iPSC-derived MNs corrected the NF subunit proportion, mitigating NF aggregation and neurite degeneration. Thus, NF misregulation underlies mutant SOD1-mediated NF aggregation and axonal degeneration in ALS MNs.
Copyright © 2014 Elsevier Inc. All rights reserved.
Figures
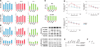
References
-
- Ban H, Nishishita N, Fusaki N, Tabata T, Saeki K, Shikamura M, Takada N, Inoue M, Hasegawa M, Kawamata S, Nishikawa S. Efficient generation of transgene-free human induced pluripotent stem cells (iPSCs) by temperature-sensitive Sendai virus vectors. Proc. Natl. Acad. Sci. USA. 2011;108:14234–14239. - PMC - PubMed
-
- Bergeron C, Beric-Maskarel K, Muntasser S, Weyer L, Somerville MJ, Percy ME. Neurofilament light and polyadenylated mRNA levels are decreased in amyotrophic lateral sclerosis motor neurons. J. Neuropathol. Exp. Neurol. 1994;53:221–230. - PubMed
-
- Bruijn LI, Houseweart MK, Kato S, Anderson KL, Anderson SD, Ohama E, Reaume AG, Scott RW, Cleveland DW. Aggregation and motor neuron toxicity of an ALS-linked SOD1 mutant independent from wild-type SOD1. Science. 1998;281:1851–1854. - PubMed
-
- Carpenter S. Proximal axonal enlargement in motor neuron disease. Neurology. 1968;18:841–851. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

