Large-scale identification and analysis of suppressive drug interactions
- PMID: 24704506
- PMCID: PMC4281482
- DOI: 10.1016/j.chembiol.2014.02.012
Large-scale identification and analysis of suppressive drug interactions
Abstract
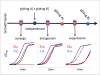
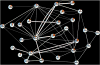
One drug may suppress the effects of another. Although knowledge of drug suppression is vital to avoid efficacy-reducing drug interactions or discover countermeasures for chemical toxins, drug-drug suppression relationships have not been systematically mapped. Here, we analyze the growth response of Saccharomyces cerevisiae to anti-fungal compound ("drug") pairs. Among 440 ordered drug pairs, we identified 94 suppressive drug interactions. Using only pairs not selected on the basis of their suppression behavior, we provide an estimate of the prevalence of suppressive interactions between anti-fungal compounds as 17%. Analysis of the drug suppression network suggested that Bromopyruvate is a frequently suppressive drug and Staurosporine is a frequently suppressed drug. We investigated potential explanations for suppressive drug interactions, including chemogenomic analysis, coaggregation, and pH effects, allowing us to explain the interaction tendencies of Bromopyruvate.
Copyright © 2014 Elsevier Ltd. All rights reserved.
Figures




Comment in
-
Suppressive drug interactions between antifungals.Chem Biol. 2014 Apr 24;21(4):439-440. doi: 10.1016/j.chembiol.2014.04.004. Chem Biol. 2014. PMID: 24766845
References
-
- Abe F, Usui K, Hiraki T. Fluconazole modulates membrane rigidity, heterogeneity, and water penetration into the plasma membrane in Saccharomyces cerevisiae. Biochemistry. 2009;48:8494–8504. - PubMed
-
- Aitken A, Learmonth M. Carboxymethylation of cysteine using iodoacetamide/iodoacetic acid. In: Walker JM, editor. The Protein Protocols Handbook. Totowa, NJ: Humana Press; 1996. pp. 339–340.
-
- Bandy J, Milward D, McQuay S. Mining protein-protein interactions from published literature using Linguamatics I2E. Methods Mol Biol. 2009;563:3–13. - PubMed
-
- Berg JM, Tymoczko JL, Stryer L. Biochemistry. New York: W.H. Freeman & Company; 2010.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

