Mechanism of activation of bacterial cellulose synthase by cyclic di-GMP
- PMID: 24704788
- PMCID: PMC4013215
- DOI: 10.1038/nsmb.2803
Mechanism of activation of bacterial cellulose synthase by cyclic di-GMP
Abstract
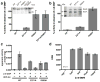
The bacterial signaling molecule cyclic di-GMP (c-di-GMP) stimulates the synthesis of bacterial cellulose, which is frequently found in biofilms. Bacterial cellulose is synthesized and translocated across the inner membrane by a complex of cellulose synthase BcsA and BcsB subunits. Here we present crystal structures of the c-di-GMP-activated BcsA-BcsB complex. The structures reveal that c-di-GMP releases an autoinhibited state of the enzyme by breaking a salt bridge that otherwise tethers a conserved gating loop that controls access to and substrate coordination at the active site. Disrupting the salt bridge by mutagenesis generates a constitutively active cellulose synthase. Additionally, the c-di-GMP-activated BcsA-BcsB complex contains a nascent cellulose polymer whose terminal glucose unit rests at a new location above BcsA's active site and is positioned for catalysis. Our mechanistic insights indicate how c-di-GMP allosterically modulates enzymatic functions.
Figures
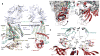



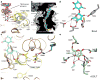
Similar articles
-
BcsA and BcsB form the catalytically active core of bacterial cellulose synthase sufficient for in vitro cellulose synthesis.Proc Natl Acad Sci U S A. 2013 Oct 29;110(44):17856-61. doi: 10.1073/pnas.1314063110. Epub 2013 Oct 14. Proc Natl Acad Sci U S A. 2013. PMID: 24127606 Free PMC article.
-
Observing cellulose biosynthesis and membrane translocation in crystallo.Nature. 2016 Mar 17;531(7594):329-34. doi: 10.1038/nature16966. Epub 2016 Mar 9. Nature. 2016. PMID: 26958837 Free PMC article.
-
Crystallographic snapshot of cellulose synthesis and membrane translocation.Nature. 2013 Jan 10;493(7431):181-6. doi: 10.1038/nature11744. Epub 2012 Dec 9. Nature. 2013. PMID: 23222542 Free PMC article.
-
Weaving of bacterial cellulose by the Bcs secretion systems.FEMS Microbiol Rev. 2022 Mar 3;46(2):fuab051. doi: 10.1093/femsre/fuab051. FEMS Microbiol Rev. 2022. PMID: 34634120 Free PMC article. Review.
-
The HD-GYP domain, cyclic di-GMP signaling, and bacterial virulence to plants.Mol Plant Microbe Interact. 2006 Dec;19(12):1378-84. doi: 10.1094/MPMI-19-1378. Mol Plant Microbe Interact. 2006. PMID: 17153922 Review.
Cited by
-
Structure of Arabidopsis CESA3 catalytic domain with its substrate UDP-glucose provides insight into the mechanism of cellulose synthesis.Proc Natl Acad Sci U S A. 2021 Mar 16;118(11):e2024015118. doi: 10.1073/pnas.2024015118. Proc Natl Acad Sci U S A. 2021. PMID: 33729990 Free PMC article.
-
Diversity of Cyclic Di-GMP-Binding Proteins and Mechanisms.J Bacteriol. 2016 Jan 1;198(1):32-46. doi: 10.1128/JB.00333-15. J Bacteriol. 2016. PMID: 26055114 Free PMC article. Review.
-
The Biology of the Escherichia coli Extracellular Matrix.Microbiol Spectr. 2015 Jun;3(3):10.1128/microbiolspec.MB-0014-2014. doi: 10.1128/microbiolspec.MB-0014-2014. Microbiol Spectr. 2015. PMID: 26185090 Free PMC article.
-
The role of RNA regulators, quorum sensing and c-di-GMP in bacterial biofilm formation.FEBS Open Bio. 2023 Jun;13(6):975-991. doi: 10.1002/2211-5463.13389. Epub 2022 Mar 13. FEBS Open Bio. 2023. PMID: 35234364 Free PMC article. Review.
-
Structural Conservation and Diversity of PilZ-Related Domains.J Bacteriol. 2020 Jan 29;202(4):e00664-19. doi: 10.1128/JB.00664-19. Print 2020 Jan 29. J Bacteriol. 2020. PMID: 31740493 Free PMC article.
References
-
- Gloag ES, Turnbull L, Huang A, Vallotton P, Wang H, Nolan LM, Mililli L, Hunt C, Lu J, Osvath SR, Monahan LG, Cavaliere R, Charles IG, Wand MP, Gee ML, Prabhakar R, Whitchurch CB. Self-organization of bacterial biofilms is facilitated by extracellular DNA. Proc Natl Acad Sci U S A. 2013;110:11541–11546. - PMC - PubMed
-
- Stewart PS, Costerton JW. Antibiotic resistance of bacteria in biofilms. Lancet. 2001;358:135–138. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

