A Gene-By-Gene Approach to Bacterial Population Genomics: Whole Genome MLST of Campylobacter
- PMID: 24704917
- PMCID: PMC3902793
- DOI: 10.3390/genes3020261
A Gene-By-Gene Approach to Bacterial Population Genomics: Whole Genome MLST of Campylobacter
Abstract
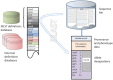
Campylobacteriosis remains a major human public health problem world-wide. Genetic analyses of Campylobacter isolates, and particularly molecular epidemiology, have been central to the study of this disease, particularly the characterization of Campylobacter genotypes isolated from human infection, farm animals, and retail food. These studies have demonstrated that Campylobacter populations are highly structured, with distinct genotypes associated with particular wild or domestic animal sources, and that chicken meat is the most likely source of most human infection in countries such as the UK. The availability of multiple whole genome sequences from Campylobacter isolates presents the prospect of identifying those genes or allelic variants responsible for host-association and increased human disease risk, but the diversity of Campylobacter genomes present challenges for such analyses. We present a gene-by-gene approach for investigating the genetic basis of phenotypes in diverse bacteria such as Campylobacter, implemented with the BIGSdb software on the pubMLST.org/campylobacter website.
Figures

References
-
- Newell D.G., Koopmans M., Verhoef L., Duizer E., Aidara-Kane A., Sprong H., Opsteegh M., Langelaar M., Threfall J., Scheutz F., et al. Food-borne diseases—The challenges of 20 years ago still persist while new ones continue to emerge. Int. J. Food Microbiol. 2010;139:S3–S15. doi: 10.1016/j.ijfoodmicro.2010.01.021. - DOI - PMC - PubMed
-
- Strachan N.J.C., Forbes K.J. The growing UK epidemic of human campylobacteriosis. Lancet. 2010;376:665–667. - PubMed
-
- Nachamkin I. Chronic effects of Campylobacter infection. Microbes Infect. 2002;4:399–403. - PubMed
-
- Young K.T., Davis L.M., Dirita V.J. Campylobacter jejuni: Molecular biology and pathogenesis. Nat. Rev. Microbiol. 2007;5:665–679. - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

