Parallel clustering algorithm for large-scale biological data sets
- PMID: 24705246
- PMCID: PMC3976248
- DOI: 10.1371/journal.pone.0091315
Parallel clustering algorithm for large-scale biological data sets
Abstract
Backgrounds: Recent explosion of biological data brings a great challenge for the traditional clustering algorithms. With increasing scale of data sets, much larger memory and longer runtime are required for the cluster identification problems. The affinity propagation algorithm outperforms many other classical clustering algorithms and is widely applied into the biological researches. However, the time and space complexity become a great bottleneck when handling the large-scale data sets. Moreover, the similarity matrix, whose constructing procedure takes long runtime, is required before running the affinity propagation algorithm, since the algorithm clusters data sets based on the similarities between data pairs.
Methods: Two types of parallel architectures are proposed in this paper to accelerate the similarity matrix constructing procedure and the affinity propagation algorithm. The memory-shared architecture is used to construct the similarity matrix, and the distributed system is taken for the affinity propagation algorithm, because of its large memory size and great computing capacity. An appropriate way of data partition and reduction is designed in our method, in order to minimize the global communication cost among processes.
Result: A speedup of 100 is gained with 128 cores. The runtime is reduced from serval hours to a few seconds, which indicates that parallel algorithm is capable of handling large-scale data sets effectively. The parallel affinity propagation also achieves a good performance when clustering large-scale gene data (microarray) and detecting families in large protein superfamilies.
Conflict of interest statement
Figures
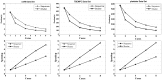
 axis represents the number of cores, and the
axis represents the number of cores, and the  axis represents the runtime and speedup.
axis represents the runtime and speedup.
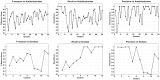
 axis and
axis and  axis represent the cluster and its corresponding best score value, respectively. Some tiny and singleton clusters are not considered in the figure.
axis represent the cluster and its corresponding best score value, respectively. Some tiny and singleton clusters are not considered in the figure.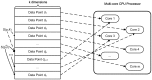
 and
and  represent the row number of the input matrix and the dimension of each row, respectively;
represent the row number of the input matrix and the dimension of each row, respectively;  represents the similarity between data point
represents the similarity between data point  and
and  ;
;  represents the number of available cores).
represents the number of available cores).

 rows and a computing cluster with
rows and a computing cluster with  machine nodes, each node is assigned about
machine nodes, each node is assigned about  rows of each information matrix. In each node, the
rows of each information matrix. In each node, the  rows are processed by
rows are processed by  cores concurrently.
cores concurrently.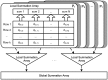
 processes
processes  , and each process is assigned about
, and each process is assigned about  rows of the availability messages matrix after partitioning. In each row, there are
rows of the availability messages matrix after partitioning. In each row, there are  column. In order to reduce the communication cost, each process computes the local summation of the
column. In order to reduce the communication cost, each process computes the local summation of the  columns and stores the intermediate values in the Local Summation Array firstly, and then these local values in all processes are gathered and scattered to compute the global summation of the
columns and stores the intermediate values in the Local Summation Array firstly, and then these local values in all processes are gathered and scattered to compute the global summation of the  columns.
columns.Similar articles
-
Parallel clustering algorithm for large data sets with applications in bioinformatics.IEEE/ACM Trans Comput Biol Bioinform. 2009 Apr-Jun;6(2):344-52. doi: 10.1109/TCBB.2007.70272. IEEE/ACM Trans Comput Biol Bioinform. 2009. PMID: 19407357
-
Clustering huge protein sequence sets in linear time.Nat Commun. 2018 Jun 29;9(1):2542. doi: 10.1038/s41467-018-04964-5. Nat Commun. 2018. PMID: 29959318 Free PMC article.
-
Markov clustering versus affinity propagation for the partitioning of protein interaction graphs.BMC Bioinformatics. 2009 Mar 30;10:99. doi: 10.1186/1471-2105-10-99. BMC Bioinformatics. 2009. PMID: 19331680 Free PMC article.
-
Methods for evaluating clustering algorithms for gene expression data using a reference set of functional classes.BMC Bioinformatics. 2006 Aug 31;7:397. doi: 10.1186/1471-2105-7-397. BMC Bioinformatics. 2006. PMID: 16945146 Free PMC article.
-
Tight clustering for large datasets with an application to gene expression data.Sci Rep. 2019 Feb 28;9(1):3053. doi: 10.1038/s41598-019-39459-w. Sci Rep. 2019. PMID: 30816195 Free PMC article.
Cited by
-
Machine learning for biomedical literature triage.PLoS One. 2014 Dec 31;9(12):e115892. doi: 10.1371/journal.pone.0115892. eCollection 2014. PLoS One. 2014. PMID: 25551575 Free PMC article.
-
A comprehensive review of machine learning algorithms and their application in geriatric medicine: present and future.Aging Clin Exp Res. 2023 Nov;35(11):2363-2397. doi: 10.1007/s40520-023-02552-2. Epub 2023 Sep 8. Aging Clin Exp Res. 2023. PMID: 37682491 Free PMC article. Review.
-
Applications of Community Detection Algorithms to Large Biological Datasets.Methods Mol Biol. 2021;2243:59-80. doi: 10.1007/978-1-0716-1103-6_3. Methods Mol Biol. 2021. PMID: 33606252
-
paraGSEA: a scalable approach for large-scale gene expression profiling.Nucleic Acids Res. 2017 Sep 29;45(17):e155. doi: 10.1093/nar/gkx679. Nucleic Acids Res. 2017. PMID: 28973463 Free PMC article.
-
Similarity measure and domain adaptation in multiple mixture model clustering: An application to image processing.PLoS One. 2017 Jul 7;12(7):e0180307. doi: 10.1371/journal.pone.0180307. eCollection 2017. PLoS One. 2017. PMID: 28686634 Free PMC article.
References
-
- Hanisch D, Zien A, Zimmer R, Lengauer T (2002) Co-clustering of biological networks and gene expression data. Bioinformatics 18: S145–S154. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

