Mechanisms of base substitution mutagenesis in cancer genomes
- PMID: 24705290
- PMCID: PMC3978516
- DOI: 10.3390/genes5010108
Mechanisms of base substitution mutagenesis in cancer genomes
Abstract
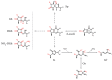
Cancer genome sequence data provide an invaluable resource for inferring the key mechanisms by which mutations arise in cancer cells, favoring their survival, proliferation and invasiveness. Here we examine recent advances in understanding the molecular mechanisms responsible for the predominant type of genetic alteration found in cancer cells, somatic single base substitutions (SBSs). Cytosine methylation, demethylation and deamination, charge transfer reactions in DNA, DNA replication timing, chromatin status and altered DNA proofreading activities are all now known to contribute to the mechanisms leading to base substitution mutagenesis. We review current hypotheses as to the major processes that give rise to SBSs and evaluate their relative relevance in the light of knowledge acquired from cancer genome sequencing projects and the study of base modifications, DNA repair and lesion bypass. Although gene expression data on APOBEC3B enzymes provide support for a role in cancer mutagenesis through U:G mismatch intermediates, the enzyme preference for single-stranded DNA may limit its activity genome-wide. For SBSs at both CG:CG and YC:GR sites, we outline evidence for a prominent role of damage by charge transfer reactions that follow interactions of the DNA with reactive oxygen species (ROS) and other endogenous or exogenous electron-abstracting molecules.
Figures





References
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

