Tbx1 is required autonomously for cell survival and fate in the pharyngeal core mesoderm to form the muscles of mastication
- PMID: 24705356
- PMCID: PMC4103673
- DOI: 10.1093/hmg/ddu140
Tbx1 is required autonomously for cell survival and fate in the pharyngeal core mesoderm to form the muscles of mastication
Abstract
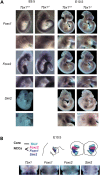
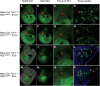
Velo-cardio-facial/DiGeorge syndrome, also known as 22q11.2 deletion syndrome, is a congenital anomaly disorder characterized by craniofacial anomalies including velo-pharyngeal insufficiency, facial muscle hypotonia and feeding difficulties, in part due to hypoplasia of the branchiomeric muscles. Inactivation of both alleles of mouse Tbx1, encoding a T-box transcription factor, deleted on chromosome 22q11.2, results in reduction or loss of branchiomeric muscles. To identify downstream pathways, we performed gene profiling of microdissected pharyngeal arch one (PA1) from Tbx1(+/+) and Tbx1(-/-) embryos at stages E9.5 (somites 20-25) and E10.5 (somites 30-35). Basic helix-loop-helix (bHLH) transcription factors were reduced, while secondary heart field genes were increased in expression early and were replaced by an increase in expression of cellular stress response genes later, suggesting a change in gene expression patterns or cell populations. Lineage tracing studies using Mesp1(Cre) and T-Cre drivers showed that core mesoderm cells within PA1 were present at E9.5 but were greatly reduced by E10.5 in Tbx1(-/-) embryos. Using Tbx1(Cre) knock-in mice, we found that cells are lost due to apoptosis, consistent with increase in expression of cellular stress response genes at E10.5. To determine whether Tbx1 is required autonomously in the core mesoderm, we used Mesp1(Cre) and T-Cre mesodermal drivers in combination with inactivate Tbx1 and found reduction or loss of branchiomeric muscles from PA1. These mechanistic studies inform us that Tbx1 is required upstream of key myogenic genes needed for core mesoderm cell survival and fate, between E9.5 and E10.5, resulting in formation of the branchiomeric muscles.
© The Author 2014. Published by Oxford University Press. All rights reserved. For Permissions, please email: journals.permissions@oup.com.
Figures



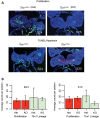
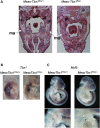
References
-
- Bryson-Richardson R.J., Currie P.D. The genetics of vertebrate myogenesis. Nat. Rev. Genet. 2008;9:632–646. - PubMed
-
- Zim S., Schelper R., Kellman R., Tatum S., Ploutz-Snyder R., Shprintzen R. Thickness and histologic and histochemical properties of the superior pharyngeal constrictor muscle in velocardiofacial syndrome. Arch. Facial Plast. Surg. 2003;5:503–510. - PubMed
-
- Lypka M., Bidros R., Rizvi M., Gaon M., Rubenstein A., Fox D., Cronin E. Posterior pharyngeal augmentation in the treatment of velopharyngeal insufficiency: a 40-year experience. Ann. Plast. Surg. 2010;65:48–51. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

