The unfolded protein response affects readthrough of premature termination codons
- PMID: 24705877
- PMCID: PMC4023889
- DOI: 10.1002/emmm.201303347
The unfolded protein response affects readthrough of premature termination codons
Abstract
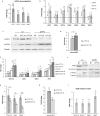
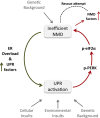
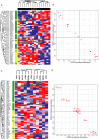
One-third of monogenic inherited diseases result from premature termination codons (PTCs). Readthrough of in-frame PTCs enables synthesis of full-length functional proteins. However, extended variability in the response to readthrough treatment is found among patients, which correlates with the level of nonsense transcripts. Here, we aimed to reveal cellular pathways affecting this inter-patient variability. We show that activation of the unfolded protein response (UPR) governs the response to readthrough treatment by regulating the levels of transcripts carrying PTCs. Quantitative proteomic analyses showed substantial differences in UPR activation between patients carrying PTCs, correlating with their response. We further found a significant inverse correlation between the UPR and nonsense-mediated mRNA decay (NMD), suggesting a feedback loop between these homeostatic pathways. We uncovered and characterized the mechanism underlying this NMD-UPR feedback loop, which augments both UPR activation and NMD attenuation. Importantly, this feedback loop enhances the response to readthrough treatment, highlighting its clinical importance. Altogether, our study demonstrates the importance of the UPR and its regulatory network for genetic diseases caused by PTCs and for cell homeostasis under normal conditions.
Figures


References
-
- Ahnesorg P, Smith P, Jackson SP. XLF interacts with the XRCC4-DNA ligase IV complex to promote DNA nonhomologous end-joining. Cell. 2006;124:301–313. - PubMed
-
- Austin RC. The unfolded protein response in health and disease. Antioxid Redox Signal. 2009;11:2279–2287. - PubMed
-
- Bartoszewski R, Rab A, Twitty G, Stevenson L, Fortenberry J, Piotrowski A, Dumanski JP, Bebok Z. The mechanism of cystic fibrosis transmembrane conductance regulator transcriptional repression during the unfolded protein response. J Biol Chem. 2008;283:12154–12165. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

