Antiretroviral therapy, interferon sensitivity, and virologic setpoint in human immunodeficiency virus/hepatitis C virus coinfected patients
- PMID: 24706559
- PMCID: PMC4110185
- DOI: 10.1002/hep.27158
Antiretroviral therapy, interferon sensitivity, and virologic setpoint in human immunodeficiency virus/hepatitis C virus coinfected patients
Abstract
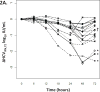
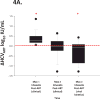
Human immunodeficiency virus (HIV) and hepatitis C virus (HCV) cause substantial mortality, especially in persons chronically infected with both viruses. HIV infection raises plasma HCV RNA levels and diminishes the response to exogenous alpha interferon (IFN). The degree to which antiretroviral therapy (ART) control of infection overcomes these HIV effects is unknown. Participants with HIV-HCV coinfection were enrolled in a trial to measure HCV viral kinetics after IFN administration (ΔHCVIFN ) twice: initially before (pre-ART) and then after (post-ART) HIV RNA suppression. Liver tissue was obtained 2-4 hours before each IFN injection to measure interferon stimulated genes (ISGs). Following ART, the ΔHCVIFN at 72 hours (ΔHCVIFN,72 ) increased in 15/19 (78.9%) participants by a median (interquartile range [IQR]) of 0.11 log10 IU/mL (0.00-0.40; P < 0.05). Increases in ΔHCVIFN,72 post-ART were associated with decreased hepatic expression of several ISGs (r = -0.68; P = 0.001); a 2-fold reduction in a four-gene ISG signature predicted an increase in ΔHCVIFN,72 of 0.78 log10 IU/mL (95% confidence interval [CI] 0.36,1.20). Pre- and post-ART ΔHCVIFN,72 were closely associated (r = 0.87; P < 0.001). HCV virologic setpoint also changed after ART (ΔHCVART ): transient median increases of 0.28 log10 IU/mL were followed by eventual median decreases from baseline of 0.21 log10 IU/mL (P = 0.002). A bivariate model of HIV RNA control (P < 0.05) and increased expression of a nine-gene ISG signature (P < 0.001) predicted the eventual decreased ΔHCVART .
Conclusion: ART is associated with lower post-IFN HCV RNA levels and that change is linked to reduced hepatic ISG expression. These data support recommendations to provide ART prior to IFN-based treatment of HCV and may provide insights into the pathogenesis of HIV-HCV coinfection.
© 2014 by the American Association for the Study of Liver Diseases.
Figures










Similar articles
-
A case of successful hepatitis C virus eradication by 24 weeks of telaprevir-based triple therapy for a hemophilia patient with hepatitis C virus/human immunodeficiency virus co-infection who previously failed pegylated interferon-α and ribavirin therapy.J Infect Chemother. 2014 May;20(5):320-4. doi: 10.1016/j.jiac.2013.11.006. Epub 2014 Jan 27. J Infect Chemother. 2014. PMID: 24477330
-
Comparative efficacy, pharmacokinetic, pharmacodynamic activity, and interferon stimulated gene expression of different interferon formulations in HIV/HCV genotype-1 infected patients.J Med Virol. 2014 Feb;86(2):177-85. doi: 10.1002/jmv.23773. Epub 2013 Oct 26. J Med Virol. 2014. PMID: 24166150
-
Low Efficacy of Pegylated Interferon plus Ribavirin plus Nitazoxanide for HCV Genotype 4 and HIV Coinfection.PLoS One. 2015 Dec 7;10(12):e0143492. doi: 10.1371/journal.pone.0143492. eCollection 2015. PLoS One. 2015. PMID: 26640956 Free PMC article. Clinical Trial.
-
Interferon combination therapy for HIV/hepatitis C virus coinfection.Immunotherapy. 2011 Sep;3(9):1087-102. doi: 10.2217/imt.11.105. Immunotherapy. 2011. PMID: 21913831 Review.
-
Recent advances in the treatment of HIV/HBV and HIV/HCV co-infection.Mini Rev Med Chem. 2012 Aug;12(9):890-904. doi: 10.2174/138955712800959062. Mini Rev Med Chem. 2012. PMID: 22530578 Review.
Cited by
-
Presence of Human Hepegivirus-1 in a Cohort of People Who Inject Drugs.Ann Intern Med. 2017 Jul 4;167(1):1-7. doi: 10.7326/M17-0085. Epub 2017 Jun 6. Ann Intern Med. 2017. PMID: 28586923 Free PMC article.
-
People with HIV-1 demonstrate type 1 interferon refractoriness associated with upregulated USP18.J Virol. 2021 Apr 26;95(10):e01777-20. doi: 10.1128/JVI.01777-20. Epub 2021 Mar 3. J Virol. 2021. PMID: 33658340 Free PMC article.
-
Intracellular HIV-1 RNA and CD4+ T-cell activation in patients starting antiretrovirals.AIDS. 2017 Jun 19;31(10):1405-1414. doi: 10.1097/QAD.0000000000001480. AIDS. 2017. PMID: 28358734 Free PMC article.
-
HIV influences clustering and intracellular replication of hepatitis C virus.J Viral Hepat. 2021 Feb;28(2):334-344. doi: 10.1111/jvh.13429. Epub 2020 Nov 24. J Viral Hepat. 2021. PMID: 33128322 Free PMC article.
-
CMPK2 and BCL-G are associated with type 1 interferon-induced HIV restriction in humans.Sci Adv. 2018 Aug 1;4(8):eaat0843. doi: 10.1126/sciadv.aat0843. eCollection 2018 Aug. Sci Adv. 2018. PMID: 30083606 Free PMC article.
References
-
- Weber R, Sabin CA, Friis-Moller N, Reiss P, El-Sadr WM, Kirk O, Dabis F, et al. Liver-related deaths in persons infected with the human immunodeficiency virus: the D:A:D study. Arch Intern Med. 2006;166:1632–1641. - PubMed
-
- Sherman KE, Rouster SD, Chung RT, Rajicic N. Hepatitis C Virus prevalence among patients infected with Human Immunodeficiency Virus: a cross-sectional analysis of the US adult AIDS Clinical Trials Group. Clin Infect Dis. 2002;34:831–837. - PubMed
-
- Thomas DL, Shih JW, Alter HJ, Vlahov D, Cohn S, Hoover DR, Cheung L, et al. Effect of human immunodeficiency virus on hepatitis C virus infection among injecting drug users. Journal of Infectious Diseases. 1996;174:690–695. - PubMed
-
- Goedert JJ, Eyster ME, Lederman MM, Mandalaki T, De Moerloose P, White GC, 2nd, Angiolillo AL, et al. End-stage liver disease in persons with hemophilia and transfusion-associated infections. Blood. 2002;100:1584–1589. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
