Role of norEF in denitrification, elucidated by physiological experiments with Rhodobacter sphaeroides
- PMID: 24706737
- PMCID: PMC4054195
- DOI: 10.1128/JB.00003-14
Role of norEF in denitrification, elucidated by physiological experiments with Rhodobacter sphaeroides
Abstract
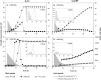
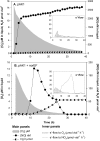
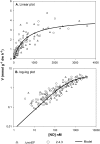
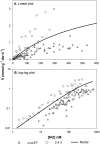
Many denitrifying organisms contain the norEF gene cluster, which codes for two proteins that are thought to be involved in denitrification because they are expressed during the reduction of nitrite and nitric oxide. The products of both genes are predicted to be membrane associated, and the norE product is a member of the cytochrome c oxidase subunit III family. However, the specific role of norEF is unknown. The denitrification phenotypes of Rhodobacter sphaeroides strains with and without norEF genes were studied, and it was found that loss of norEF lowered the rate of denitrification from nitrate and resulted in accumulation of micromolar concentrations of nitric oxide during denitrification from nitrite. norEF appears to have no direct role in the reduction of nitric oxide; however, since deletion of norEF in the wild-type 2.4.3 strain had essentially no influence on the kinetics of potential nitric oxide reduction (Vmax and Ks), as measured by monitoring the depletion of a bolus of nitric oxide injected into anoxic cultures without any other electron acceptors. However, norEF-deficient cells that had undergone a more chronic exposure to micromolar concentrations of nitric oxide showed an ∼50% reduction in Vmax but no change in apparent Ks. These results can explain the occurrence of norEF in the 2.4.3 strain of R. sphaeroides, which can reduce nitrate to nitrous oxide, and their absence from strains such as 2.4.1, which likely use nitric oxide reductase to mitigate stress due to episodic exposure to nitric oxide from exogenous sources.
Copyright © 2014, American Society for Microbiology. All Rights Reserved.
Figures


Similar articles
-
Identification, functional studies, and genomic comparisons of new members of the NnrR regulon in Rhodobacter sphaeroides.J Bacteriol. 2010 Feb;192(4):903-11. doi: 10.1128/JB.01026-09. Epub 2009 Dec 4. J Bacteriol. 2010. PMID: 19966004 Free PMC article.
-
Requirement of nitric oxide for induction of genes whose products are involved in nitric oxide metabolism in Rhodobacter sphaeroides 2.4.3.J Biol Chem. 1996 Oct 4;271(40):24382-8. doi: 10.1074/jbc.271.40.24382. J Biol Chem. 1996. PMID: 8798693
-
Analysis of the role of the nnrR gene product in the response of Rhodobacter sphaeroides 2.4.1 to exogenous nitric oxide.J Bacteriol. 1997 Sep;179(17):5618-20. doi: 10.1128/jb.179.17.5618-5620.1997. J Bacteriol. 1997. PMID: 9287025 Free PMC article.
-
Cell biology and molecular basis of denitrification.Microbiol Mol Biol Rev. 1997 Dec;61(4):533-616. doi: 10.1128/mmbr.61.4.533-616.1997. Microbiol Mol Biol Rev. 1997. PMID: 9409151 Free PMC article. Review.
-
Metabolic regulation including anaerobic metabolism in Paracoccus denitrificans.J Bioenerg Biomembr. 1991 Apr;23(2):163-85. doi: 10.1007/BF00762216. J Bioenerg Biomembr. 1991. PMID: 2050653 Review.
Cited by
-
Highly diverse nirK genes comprise two major clades that harbour ammonium-producing denitrifiers.BMC Genomics. 2016 Feb 29;17:155. doi: 10.1186/s12864-016-2465-0. BMC Genomics. 2016. PMID: 26923558 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

