New insights into the DT40 B cell receptor cluster using a proteomic proximity labeling assay
- PMID: 24706754
- PMCID: PMC4031500
- DOI: 10.1074/jbc.M113.529578
New insights into the DT40 B cell receptor cluster using a proteomic proximity labeling assay
Abstract
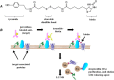
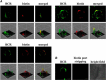
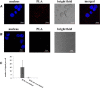
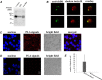
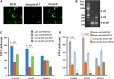
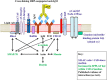
In the vertebrate immune system, each B-lymphocyte expresses a surface IgM-class B cell receptor (BCR). When cross-linked by antigen or anti-IgM antibody, the BCR accumulates with other proteins into distinct surface clusters that activate cell signaling, division, or apoptosis. However, the molecular composition of these clusters is not well defined. Here we describe a quantitative assay we call selective proteomic proximity labeling using tyramide (SPPLAT). It allows proteins in the immediate vicinity of a target to be selectively biotinylated, and hence isolated for mass spectrometry analysis. Using the chicken B cell line DT40 as a model, we use SPPLAT to provide the first proteomic analysis of any BCR cluster using proximity labeling. We detect known components of the BCR cluster, including integrins, together with proteins not previously thought to be BCR-associated. In particular, we identify the chicken B-lymphocyte allotypic marker chB6. We show that chB6 moves to within about 30-40 nm of the BCR following BCR cross-linking, and we show that cross-linking chB6 activates cell binding to integrin substrates laminin and gelatin. Our work provides new insights into the nature and composition of the BCR cluster, and confirms SPPLAT as a useful research tool in molecular and cellular proteomics.
Keywords: B Cell Receptor; Evi2a; Immunology; Integrin; Lymphocyte; Mass Spectrometry (MS); Proteomics; Raftlin; SILAC; chB6.
© 2014 by The American Society for Biochemistry and Molecular Biology, Inc.
Figures


References
Publication types
MeSH terms
Substances
Grants and funding
- BB/H024085/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- 094470/Z/10/Z/WT_/Wellcome Trust/United Kingdom
- BB/J021091/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- G0500707/MRC_/Medical Research Council/United Kingdom
- RG/09/003/27122/BHF_/British Heart Foundation/United Kingdom
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

