Proteomic analysis of Vibrio cholerae outer membrane vesicles
- PMID: 24706774
- PMCID: PMC3992640
- DOI: 10.1073/pnas.1403683111
Proteomic analysis of Vibrio cholerae outer membrane vesicles
Abstract
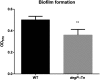
Outer membrane vesicles (OMVs) produced by Gram-negative bacteria provide an interesting research material for defining cell-envelope proteins without experimental cell disruption. OMVs are also promising immunogenic platforms and may play important roles in bacterial survival and pathogenesis. We used in-solution trypsin digestion coupled to mass spectrometry to identify 90 proteins present in OMVs of Vibrio cholerae when grown under conditions that activate the TCP pilus virulence regulatory protein (ToxT) virulence regulon. The ToxT expression profile and potential contribution to virulence of these proteins were assessed using ToxT and in vivo RNA-seq, Tn-seq, and cholera stool proteomic and other genome-wide data sets. Thirteen OMV-associated proteins appear to be essential for cell growth, and therefore may represent antibacterial drug targets. Another 12 nonessential OMV proteins, including DegP protease, were required for intestinal colonization in rabbits. Comparative proteomics of a degP mutant revealed the importance of DegP in the incorporation of nine proteins into OMVs, including ones involved in biofilm matrix formation and various substrates of the type II secretion system. Taken together, these results suggest that DegP plays an important role in determining the content of OMVs and also affects phenotypes such as intestinal colonization, proper function of the type II secretion system, and formation of biofilm matrix.
Keywords: CTX phage; HtrA family; biofilm formation; in-solution digestion.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
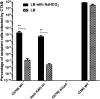
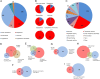
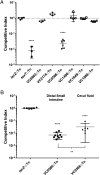
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

