Functional characterization of flavobacteria rhodopsins reveals a unique class of light-driven chloride pump in bacteria
- PMID: 24706784
- PMCID: PMC4020065
- DOI: 10.1073/pnas.1403051111
Functional characterization of flavobacteria rhodopsins reveals a unique class of light-driven chloride pump in bacteria
Abstract
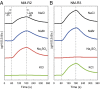
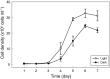
Light-activated, ion-pumping rhodopsins are broadly distributed among many different bacteria and archaea inhabiting the photic zone of aquatic environments. Bacterial proton- or sodium-translocating rhodopsins can convert light energy into a chemiosmotic force that can be converted into cellular biochemical energy, and thus represent a widespread alternative form of photoheterotrophy. Here we report that the genome of the marine flavobacterium Nonlabens marinus S1-08(T) encodes three different types of rhodopsins: Nonlabens marinus rhodopsin 1 (NM-R1), Nonlabens marinus rhodopsin 2 (NM-R2), and Nonlabens marinus rhodopsin 3 (NM-R3). Our functional analysis demonstrated that NM-R1 and NM-R2 are light-driven outward-translocating H(+) and Na(+) pumps, respectively. Functional analyses further revealed that the light-activated NM-R3 rhodopsin pumps Cl(-) ions into the cell, representing the first chloride-pumping rhodopsin uncovered in a marine bacterium. Phylogenetic analysis revealed that NM-R3 belongs to a distinct phylogenetic lineage quite distant from archaeal inward Cl(-)-pumping rhodopsins like halorhodopsin, suggesting that different types of chloride-pumping rhodopsins have evolved independently within marine bacterial lineages. Taken together, our data suggest that similar to haloarchaea, a considerable variety of rhodopsin types with different ion specificities have evolved in marine bacteria, with individual marine strains containing as many as three functionally different rhodopsins.
Keywords: ecology; evolution; photoheterotroph; photoproteins; retinal.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


Comment in
-
Nature's toolkit for microbial rhodopsin ion pumps.Proc Natl Acad Sci U S A. 2014 May 6;111(18):6538-9. doi: 10.1073/pnas.1405093111. Epub 2014 Apr 15. Proc Natl Acad Sci U S A. 2014. PMID: 24737891 Free PMC article. No abstract available.
References
-
- Spudich JL, Yang CS, Jung KH, Spudich EN. Retinylidene proteins: Structures and functions from archaea to humans. Annu Rev Cell Dev Biol. 2000;16:365–392. - PubMed
-
- Oesterhelt D, Stoeckenius W. Rhodopsin-like protein from the purple membrane of Halobacterium halobium. Nat New Biol. 1971;233(39):149–152. - PubMed
-
- Matsuno-Yagi A, Mukohata Y. Two possible roles of bacteriorhodopsin: A comparative study of strains of Halobacterium halobium differing in pigmentation. Biochem Biophys Res Commun. 1977;78(1):237–243. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

