VCGDB: a dynamic genome database of the Chinese population
- PMID: 24708222
- PMCID: PMC4028056
- DOI: 10.1186/1471-2164-15-265
VCGDB: a dynamic genome database of the Chinese population
Abstract
Background: The data released by the 1000 Genomes Project contain an increasing number of genome sequences from different nations and populations with a large number of genetic variations. As a result, the focus of human genome studies is changing from single and static to complex and dynamic. The currently available human reference genome (GRCh37) is based on sequencing data from 13 anonymous Caucasian volunteers, which might limit the scope of genomics, transcriptomics, epigenetics, and genome wide association studies.
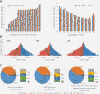
Description: We used the massive amount of sequencing data published by the 1000 Genomes Project Consortium to construct the Virtual Chinese Genome Database (VCGDB), a dynamic genome database of the Chinese population based on the whole genome sequencing data of 194 individuals. VCGDB provides dynamic genomic information, which contains 35 million single nucleotide variations (SNVs), 0.5 million insertions/deletions (indels), and 29 million rare variations, together with genomic annotation information. VCGDB also provides a highly interactive user-friendly virtual Chinese genome browser (VCGBrowser) with functions like seamless zooming and real-time searching. In addition, we have established three population-specific consensus Chinese reference genomes that are compatible with mainstream alignment software.
Conclusions: VCGDB offers a feasible strategy for processing big data to keep pace with the biological data explosion by providing a robust resource for genomics studies; in particular, studies aimed at finding regions of the genome associated with diseases.
Figures


Similar articles
-
[Database resources of the reference genome and genetic variation maps for the Chinese population].Yi Chuan. 2018 Nov 20;40(11):1048-1054. doi: 10.16288/j.yczz.18-177. Yi Chuan. 2018. PMID: 30465539 Chinese.
-
Whole Genome Analyses of Chinese Population and De Novo Assembly of A Northern Han Genome.Genomics Proteomics Bioinformatics. 2019 Jun;17(3):229-247. doi: 10.1016/j.gpb.2019.07.002. Epub 2019 Sep 5. Genomics Proteomics Bioinformatics. 2019. PMID: 31494266 Free PMC article.
-
PGG.Han: the Han Chinese genome database and analysis platform.Nucleic Acids Res. 2020 Jan 8;48(D1):D971-D976. doi: 10.1093/nar/gkz829. Nucleic Acids Res. 2020. PMID: 31584086 Free PMC article.
-
Species-Specific Genome Sequence Databases: A Practical Review.Methods Mol Biol. 2017;1533:173-181. doi: 10.1007/978-1-4939-6658-5_9. Methods Mol Biol. 2017. PMID: 27987170 Review.
-
The evolution of dbSNP: 25 years of impact in genomic research.Nucleic Acids Res. 2025 Jan 6;53(D1):D925-D931. doi: 10.1093/nar/gkae977. Nucleic Acids Res. 2025. PMID: 39530225 Free PMC article. Review.
Cited by
-
T2T-YAO, T2T-SHUN, and more.Genomics Proteomics Bioinformatics. 2023 Dec;21(6):1081-1082. doi: 10.1016/j.gpb.2023.09.002. Epub 2023 Sep 22. Genomics Proteomics Bioinformatics. 2023. PMID: 37742994 Free PMC article. No abstract available.
-
SATB2-associated syndrome caused by a novel SATB2 mutation in a Chinese boy: A case report and literature review.World J Clin Cases. 2021 Jul 26;9(21):6081-6090. doi: 10.12998/wjcc.v9.i21.6081. World J Clin Cases. 2021. PMID: 34368330 Free PMC article.
-
The BIG Data Center: from deposition to integration to translation.Nucleic Acids Res. 2017 Jan 4;45(D1):D18-D24. doi: 10.1093/nar/gkw1060. Epub 2016 Nov 28. Nucleic Acids Res. 2017. PMID: 27899658 Free PMC article.
-
Effects of Polymorphisms in APOA4-APOA5-ZNF259-BUD13 Gene Cluster on Plasma Levels of Triglycerides and Risk of Coronary Heart Disease in a Chinese Han Population.PLoS One. 2015 Sep 23;10(9):e0138652. doi: 10.1371/journal.pone.0138652. eCollection 2015. PLoS One. 2015. PMID: 26397108 Free PMC article.
-
RGAAT: A Reference-based Genome Assembly and Annotation Tool for New Genomes and Upgrade of Known Genomes.Genomics Proteomics Bioinformatics. 2018 Oct;16(5):373-381. doi: 10.1016/j.gpb.2018.03.006. Epub 2018 Dec 21. Genomics Proteomics Bioinformatics. 2018. PMID: 30583062 Free PMC article.
References
-
- Genetic Analysis of Psoriasis C. Strange A, Capon F, Spencer CC, Knight J, Weale ME, Allen MH, Barton A, Band G, Bellenguez C, Bergboer JG, Blackwell JM, Bramon E, Bumpstead SJ, Casas JP, Cork MJ, Corvin A, Deloukas P, Dilthey A, Duncanson A, Edkins S, Estivill X, Fitzgerald O, Freeman C, Giardina E, Gray E, Hofer A, Hüffmeier U, Hunt SE. et al.A genome-wide association study identifies new psoriasis susceptibility loci and an interaction between HLA-C and ERAP1. Nat Genet. 2010;15:985–990. doi: 10.1038/ng.694. - DOI - PMC - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

