Stringent homology-based prediction of H. sapiens-M. tuberculosis H37Rv protein-protein interactions
- PMID: 24708540
- PMCID: PMC4022245
- DOI: 10.1186/1745-6150-9-5
Stringent homology-based prediction of H. sapiens-M. tuberculosis H37Rv protein-protein interactions
Abstract
Background: H. sapiens-M. tuberculosis H37Rv protein-protein interaction (PPI) data are essential for understanding the infection mechanism of the formidable pathogen M. tuberculosis H37Rv. Computational prediction is an important strategy to fill the gap in experimental H. sapiens-M. tuberculosis H37Rv PPI data. Homology-based prediction is frequently used in predicting both intra-species and inter-species PPIs. However, some limitations are not properly resolved in several published works that predict eukaryote-prokaryote inter-species PPIs using intra-species template PPIs.
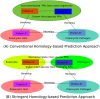
Results: We develop a stringent homology-based prediction approach by taking into account (i) differences between eukaryotic and prokaryotic proteins and (ii) differences between inter-species and intra-species PPI interfaces. We compare our stringent homology-based approach to a conventional homology-based approach for predicting host-pathogen PPIs, based on cellular compartment distribution analysis, disease gene list enrichment analysis, pathway enrichment analysis and functional category enrichment analysis. These analyses support the validity of our prediction result, and clearly show that our approach has better performance in predicting H. sapiens-M. tuberculosis H37Rv PPIs. Using our stringent homology-based approach, we have predicted a set of highly plausible H. sapiens-M. tuberculosis H37Rv PPIs which might be useful for many of related studies. Based on our analysis of the H. sapiens-M. tuberculosis H37Rv PPI network predicted by our stringent homology-based approach, we have discovered several interesting properties which are reported here for the first time. We find that both host proteins and pathogen proteins involved in the host-pathogen PPIs tend to be hubs in their own intra-species PPI network. Also, both host and pathogen proteins involved in host-pathogen PPIs tend to have longer primary sequence, tend to have more domains, tend to be more hydrophilic, etc. And the protein domains from both host and pathogen proteins involved in host-pathogen PPIs tend to have lower charge, and tend to be more hydrophilic.
Conclusions: Our stringent homology-based prediction approach provides a better strategy in predicting PPIs between eukaryotic hosts and prokaryotic pathogens than a conventional homology-based approach. The properties we have observed from the predicted H. sapiens-M. tuberculosis H37Rv PPI network are useful for understanding inter-species host-pathogen PPI networks and provide novel insights for host-pathogen interaction studies.
Figures


References
-
- Hestvik A, Hmama Z, Av-Gay Y. Mycobacterial manipulation of the host cell. FEMS Microbiol Rev. 2006;9(5):1041–1050. - PubMed
-
- Programme GT. Global Tuberculosis Control: WHO Report. Global Tuberculosis Programme: World Health Organization; 2010.
-
- Zhou H, Jin J, Wong L. Progress in computational studies of host-pathogen interactions. J Bioinform Comput Biol. 2013;9(2):26. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

