Circulating cell-free methylated DNA and lactate dehydrogenase release in colorectal cancer
- PMID: 24708595
- PMCID: PMC4021086
- DOI: 10.1186/1471-2407-14-245
Circulating cell-free methylated DNA and lactate dehydrogenase release in colorectal cancer
Abstract
Background: Hypermethylation of DNA is an epigenetic alteration commonly found in colorectal cancer (CRC) and can also be detected in blood samples of cancer patients. Methylation of the genes helicase-like transcription factor (HLTF) and hyperplastic polyposis 1 (HPP1) have been proposed as prognostic, and neurogenin 1 (NEUROG1) as diagnostic biomarker. However the underlying mechanisms leading to the release of these genes are unclear. This study aimed at examining the possible correlation of the presence of methylated genes NEUROG1, HLTF and HPP1 in serum with tissue breakdown as a possible mechanism using serum lactate dehydrogenase (LDH) as a surrogate marker. Additionally the prognostic impact of these markers was examined.
Methods: Pretherapeutic serum samples from 259 patients from all cancer stages were analyzed. Presence of hypermethylation of the genes HLTF, HPP1, and NEUROG1 was examined using methylation-specific quantitative PCR (MethyLight). LDH was determined using an UV kinetic test.
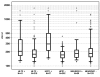
Results: Hypermethylation of HLTF and HPP1 was detected significantly more often in patients with elevated LDH levels (32% vs. 12% [p = 0.0005], and 68% vs. 11% [p < 0.0001], respectively). Also, higher LDH values correlated with a higher percentage of a fully methylated reference in a linear fashion (Spearman correlation coefficient 0.18 for HLTF [p = 0.004]; 0.49 [p < .0001] for HPP1). No correlation between methylation of NEUROG1 and LDH was found in this study. Concerning the clinical characteristics, high levels of LDH as well as methylation of HLTF and HPP1 were significantly associated with larger and more advanced stages of CRC. Accordingly, these three markers were correlated with significantly shorter survival in the overall population. Moreover, all three identified patients with a worse prognosis in the subgroup of stage IV patients.
Conclusions: We were able to provide evidence that methylation of HLTF and especially HPP1 detected in serum is strongly correlated with cell death in CRC using LDH as surrogate marker. Additionally, we found that prognostic information is given by both HLTF and HPP1 as well as LDH. In sum, determining the methylation of HLTF and HPP1 in serum might be useful in order to identify patients with more aggressive tumors.
Figures

Similar articles
-
Prognostic role of methylated free circulating DNA in colorectal cancer.Int J Cancer. 2012 Nov 15;131(10):2308-19. doi: 10.1002/ijc.27505. Epub 2012 Mar 27. Int J Cancer. 2012. PMID: 22362391
-
Methylated free-circulating HPP1 DNA is an early response marker in patients with metastatic colorectal cancer.Int J Cancer. 2017 May 1;140(9):2134-2144. doi: 10.1002/ijc.30625. Epub 2017 Feb 22. Int J Cancer. 2017. PMID: 28124380
-
Methylation of helicase-like transcription factor in serum of patients with colorectal cancer is an independent predictor of disease recurrence.Eur J Gastroenterol Hepatol. 2009 May;21(5):565-9. doi: 10.1097/MEG.0b013e328318ecf2. Eur J Gastroenterol Hepatol. 2009. PMID: 19282772
-
The helicase-like transcription factor (HLTF) in cancer: loss of function or oncomorphic conversion of a tumor suppressor?Cell Mol Life Sci. 2016 Jan;73(1):129-47. doi: 10.1007/s00018-015-2060-6. Cell Mol Life Sci. 2016. PMID: 26472339 Free PMC article. Review.
-
The helicase-like transcription factor and its implication in cancer progression.Cell Mol Life Sci. 2008 Feb;65(4):591-604. doi: 10.1007/s00018-007-7392-4. Cell Mol Life Sci. 2008. PMID: 18034322 Free PMC article. Review.
Cited by
-
Prognostic DNA methylation markers for sporadic colorectal cancer: a systematic review.Clin Epigenetics. 2018 Mar 14;10:35. doi: 10.1186/s13148-018-0461-8. eCollection 2018. Clin Epigenetics. 2018. PMID: 29564023 Free PMC article.
-
Value of Serum NEUROG1 Methylation for the Detection of Advanced Adenomas and Colorectal Cancer.Diagnostics (Basel). 2020 Jun 28;10(7):437. doi: 10.3390/diagnostics10070437. Diagnostics (Basel). 2020. PMID: 32605302 Free PMC article.
-
Nomograms based on lactate dehydrogenase to albumin ratio for predicting survival in colorectal cancer.Int J Med Sci. 2022 May 29;19(6):1003-1012. doi: 10.7150/ijms.71971. eCollection 2022. Int J Med Sci. 2022. PMID: 35813299 Free PMC article.
-
Helicase-like transcription factor (HLTF)-deleted CDX/TME model of colorectal cancer increased transcription of oxidative phosphorylation genes and diverted glycolysis to boost S-glutathionylation in lymphatic intravascular metastatic niches.PLoS One. 2023 Sep 8;18(9):e0291023. doi: 10.1371/journal.pone.0291023. eCollection 2023. PLoS One. 2023. PMID: 37682902 Free PMC article.
-
The prognostic value of lactate dehydrogenase levels in colorectal cancer: a meta-analysis.BMC Cancer. 2016 Mar 25;16:249. doi: 10.1186/s12885-016-2276-3. BMC Cancer. 2016. PMID: 27016045 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

