Patient-derived xenografts of triple-negative breast cancer reproduce molecular features of patient tumors and respond to mTOR inhibition
- PMID: 24708766
- PMCID: PMC4053092
- DOI: 10.1186/bcr3640
Patient-derived xenografts of triple-negative breast cancer reproduce molecular features of patient tumors and respond to mTOR inhibition
Abstract
Introduction: Triple-negative breast cancer (TNBC) is aggressive and lacks targeted therapies. Phosphatidylinositide 3-kinase (PI3K)/mammalian target of rapamycin (mTOR) pathways are frequently activated in TNBC patient tumors at the genome, gene expression and protein levels, and mTOR inhibitors have been shown to inhibit growth in TNBC cell lines. We describe a panel of patient-derived xenografts representing multiple TNBC subtypes and use them to test preclinical drug efficacy of two mTOR inhibitors, sirolimus (rapamycin) and temsirolimus (CCI-779).
Methods: We generated a panel of seven patient-derived orthotopic xenografts from six primary TNBC tumors and one metastasis. Patient tumors and corresponding xenografts were compared by histology, immunohistochemistry, array comparative genomic hybridization (aCGH) and phosphatidylinositol-4,5-bisphosphate 3-kinase, catalytic subunit alpha (PIK3CA) sequencing; TNBC subtypes were determined. Using a previously published logistic regression approach, we generated a rapamycin response signature from Connectivity Map gene expression data and used it to predict rapamycin sensitivity in 1,401 human breast cancers of different intrinsic subtypes, prompting in vivo testing of mTOR inhibitors and doxorubicin in our TNBC xenografts.
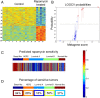
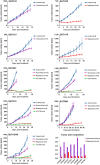
Results: Patient-derived xenografts recapitulated histology, biomarker expression and global genomic features of patient tumors. Two primary tumors had PIK3CA coding mutations, and five of six primary tumors showed flanking intron single nucleotide polymorphisms (SNPs) with conservation of sequence variations between primary tumors and xenografts, even on subsequent xenograft passages. Gene expression profiling showed that our models represent at least four of six TNBC subtypes. The rapamycin response signature predicted sensitivity for 94% of basal-like breast cancers in a large dataset. Drug testing of mTOR inhibitors in our xenografts showed 77 to 99% growth inhibition, significantly more than doxorubicin; protein phosphorylation studies indicated constitutive activation of the mTOR pathway that decreased with treatment. However, no tumor was completely eradicated.
Conclusions: A panel of patient-derived xenograft models covering a spectrum of TNBC subtypes was generated that histologically and genomically matched original patient tumors. Consistent with in silico predictions, mTOR inhibitor testing in our TNBC xenografts showed significant tumor growth inhibition in all, suggesting that mTOR inhibitors can be effective in TNBC, but will require use with additional therapies, warranting investigation of optimal drug combinations.
Figures




References
-
- Badve S, Dabbs DJ, Schnitt SJ, Baehner FL, Decker T, Eusebi V, Fox SB, Ichihara S, Jacquemier J, Lakhani SR, Palacios J, Rakha EA, Richardson AL, Schmitt FC, Tan PH, Tse GM, Weigelt B, Ellis IO, Reis-Filho JS. Basal-like and triple-negative breast cancers: a critical review with an emphasis on the implications for pathologists and oncologists. Mod Pathol. 2011;24:157–167. doi: 10.1038/modpathol.2010.200. - DOI - PubMed
-
- Kyndi M, Sørensen FB, Knudsen H, Overgaard M, Nielsen HM, Overgaard J. Danish Breast Cancer Cooperative Group. Estrogen receptor, progesterone receptor, HER-2, and response to postmastectomy radiotherapy in high-risk breast cancer: the Danish Breast Cancer Cooperative Group. J Clin Oncol. 2008;26:1419–1426. doi: 10.1200/JCO.2007.14.5565. - DOI - PubMed
-
- Perou CM, Sørlie T, Eisen MB, van de Rijn M, Jeffrey SS, Rees CA, Pollack JR, Ross DT, Johnsen H, Akslen LA, Fluge O, Pergamenschikov A, Williams C, Zhu SX, Lønning PE, Børresen-Dale AL, Brown PO, Botstein D. Molecular portraits of human breast tumours. Nature. 2000;406:747–752. doi: 10.1038/35021093. - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

