Recent advances in the structural and molecular biology of type IV secretion systems
- PMID: 24709394
- PMCID: PMC4182333
- DOI: 10.1016/j.sbi.2014.02.006
Recent advances in the structural and molecular biology of type IV secretion systems
Abstract
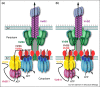
Bacteria use type IV secretion (T4S) systems to deliver DNA and protein substrates to a diverse range of prokaryotic and eukaryotic target cells. T4S systems have great impact on human health, as they are a major source of antibiotic resistance spread among bacteria and are central to infection processes of many pathogens. Therefore, deciphering the structure and underlying translocation mechanism of T4S systems is crucial to facilitate development of new drugs. The last five years have witnessed considerable progress in unraveling the structure of T4S system subassemblies, notably that of the T4S system core complex, a large 1 MegaDalton (MDa) structure embedded in the double membrane of Gram-negative bacteria and made of 3 of the 12 T4S system components. However, the recent determination of the structure of -3MDa assembly of 8 of these components has revolutionized our views of T4S system architecture and opened up new avenues of research, which are discussed in this review.
Copyright © 2014 The Authors. Published by Elsevier Ltd.. All rights reserved.
Figures



References
-
- Low H.H., Gubellini F., Rivera-Calzada A.F., Braun N., Connery S., Dujeancourt A., Lu F., Redzej A., Fronzes R., Orlova E.V., Waksman G. Structure of a type IV secretion system. Nature. 2014 doi: 10.1038/nature13081. - DOI - PMC - PubMed
-
The first structural characterization of a ∼3 MDa T4S apparatus composed of 8 of the 12 T4S system components that demonstrated the architecture of T4S machines.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

