Comparative primate genomics: emerging patterns of genome content and dynamics
- PMID: 24709753
- PMCID: PMC4113315
- DOI: 10.1038/nrg3707
Comparative primate genomics: emerging patterns of genome content and dynamics
Abstract
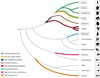
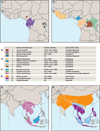
Advances in genome sequencing technologies have created new opportunities for comparative primate genomics. Genome assemblies have been published for various primate species, and analyses of several others are underway. Whole-genome assemblies for the great apes provide remarkable new information about the evolutionary origins of the human genome and the processes involved. Genomic data for macaques and other non-human primates offer valuable insights into genetic similarities and differences among species that are used as models for disease-related research. This Review summarizes current knowledge regarding primate genome content and dynamics, and proposes a series of goals for the near future.
Figures

References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

