Quantifying ChIP-seq data: a spiking method providing an internal reference for sample-to-sample normalization
- PMID: 24709819
- PMCID: PMC4079971
- DOI: 10.1101/gr.168260.113
Quantifying ChIP-seq data: a spiking method providing an internal reference for sample-to-sample normalization
Abstract
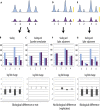
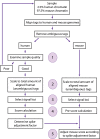
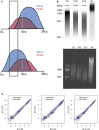
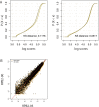
Chromatin immunoprecipitation followed by deep sequencing (ChIP-seq) experiments are widely used to determine, within entire genomes, the occupancy sites of any protein of interest, including, for example, transcription factors, RNA polymerases, or histones with or without various modifications. In addition to allowing the determination of occupancy sites within one cell type and under one condition, this method allows, in principle, the establishment and comparison of occupancy maps in various cell types, tissues, and conditions. Such comparisons require, however, that samples be normalized. Widely used normalization methods that include a quantile normalization step perform well when factor occupancy varies at a subset of sites, but may miss uniform genome-wide increases or decreases in site occupancy. We describe a spike adjustment procedure (SAP) that, unlike commonly used normalization methods intervening at the analysis stage, entails an experimental step prior to immunoprecipitation. A constant, low amount from a single batch of chromatin of a foreign genome is added to the experimental chromatin. This "spike" chromatin then serves as an internal control to which the experimental signals can be adjusted. We show that the method improves similarity between replicates and reveals biological differences including global and largely uniform changes.
© 2014 Bonhoure et al.; Published by Cold Spring Harbor Laboratory Press.
Figures


References
-
- Barski A, Cuddapah S, Cui K, Roh TY, Schones DE, Wang Z, Wei G, Chepelev I, Zhao K 2007. High-resolution profiling of histone methylations in the human genome. Cell 129: 823–837 - PubMed
-
- Chong SS, Hu P, Hernandez N 2001. Reconstitution of transcription from the human U6 small nuclear RNA promoter with eight recombinant polypeptides and a partially purified RNA polymerase III complex. J Biol Chem 276: 20727–20734 - PubMed
-
- Ciesla M, Boguta M 2008. Regulation of RNA polymerase III transcription by Maf1 protein. Acta Biochim Pol 55: 215–225 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous
