Affinity- and specificity-enhancing mutations are frequent in multispecific interactions between TIMP2 and MMPs
- PMID: 24710006
- PMCID: PMC3977929
- DOI: 10.1371/journal.pone.0093712
Affinity- and specificity-enhancing mutations are frequent in multispecific interactions between TIMP2 and MMPs
Abstract
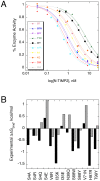
Multispecific proteins play a major role in controlling various functions such as signaling, regulation of transcription/translation, and immune response. Hence, a thorough understanding of the atomic-level principles governing multispecific interactions is important not only for the advancement of basic science but also for applied research such as drug design. Here, we study evolution of an exemplary multispecific protein, a Tissue Inhibitor of Matrix Metalloproteinases 2 (TIMP2) that binds with comparable affinities to more than twenty-six members of the Matrix Metalloproteinase (MMP) and the related ADAMs families. We postulate that due to its multispecific nature, TIMP2 is not optimized to bind to any individual MMP type, but rather embodies a compromise required for interactions with all MMPs. To explore this hypothesis, we perform computational saturation mutagenesis of the TIMP2 binding interface and predict changes in free energy of binding to eight MMP targets. Computational results reveal the non-optimality of the TIMP2 binding interface for all studied proteins, identifying many affinity-enhancing mutations at multiple positions. Several TIMP2 point mutants predicted to enhance binding affinity and/or binding specificity towards MMP14 were selected for experimental verification. Experimental results show high abundance of affinity-enhancing mutations in TIMP2, with some point mutations producing more than ten-fold improvement in affinity to MMP14. Our computational and experimental results collaboratively demonstrate that the TIMP2 sequence lies far from the fitness maximum when interacting with its target enzymes. This non-optimality of the binding interface and high potential for improvement might characterize all proteins evolved for binding to multiple targets.
Conflict of interest statement
Figures



Similar articles
-
Development of High Affinity and High Specificity Inhibitors of Matrix Metalloproteinase 14 through Computational Design and Directed Evolution.J Biol Chem. 2017 Feb 24;292(8):3481-3495. doi: 10.1074/jbc.M116.756718. Epub 2017 Jan 13. J Biol Chem. 2017. PMID: 28087697 Free PMC article.
-
Quantitative mapping of binding specificity landscapes for homologous targets by using a high-throughput method.Biochem J. 2020 May 15;477(9):1701-1719. doi: 10.1042/BCJ20200188. Biochem J. 2020. PMID: 32296833 Free PMC article.
-
Predicting affinity- and specificity-enhancing mutations at protein-protein interfaces.Biochem Soc Trans. 2013 Oct;41(5):1166-9. doi: 10.1042/BST20130121. Biochem Soc Trans. 2013. PMID: 24059503 Review.
-
Thermodynamic Basis of Selectivity in the Interactions of Tissue Inhibitors of Metalloproteinases N-domains with Matrix Metalloproteinases-1, -3, and -14.J Biol Chem. 2016 May 20;291(21):11348-58. doi: 10.1074/jbc.M116.720250. Epub 2016 Mar 31. J Biol Chem. 2016. PMID: 27033700 Free PMC article.
-
Membrane-type matrix metalloproteinases (MT-MMPs): expression and function during glioma invasion.J Neurooncol. 2001 Jun;53(2):187-202. doi: 10.1023/a:1012213604731. J Neurooncol. 2001. PMID: 11716070 Review.
Cited by
-
Climbing Up and Down Binding Landscapes through Deep Mutational Scanning of Three Homologous Protein-Protein Complexes.J Am Chem Soc. 2021 Oct 20;143(41):17261-17275. doi: 10.1021/jacs.1c08707. Epub 2021 Oct 5. J Am Chem Soc. 2021. PMID: 34609866 Free PMC article.
-
Mapping protein selectivity landscapes using multi-target selective screening and next-generation sequencing of combinatorial libraries.Nat Commun. 2018 Sep 26;9(1):3935. doi: 10.1038/s41467-018-06403-x. Nat Commun. 2018. PMID: 30258049 Free PMC article.
-
Designed Loop Extension Followed by Combinatorial Screening Confers High Specificity to a Broad Matrix MetalloproteinaseInhibitor.J Mol Biol. 2023 Jul 1;435(13):168095. doi: 10.1016/j.jmb.2023.168095. Epub 2023 Apr 15. J Mol Biol. 2023. PMID: 37068580 Free PMC article.
-
Bioorthogonal PEGylation Prolongs the Elimination Half-Life of N-TIMP2 While Retaining MMP Inhibition.Bioconjug Chem. 2022 May 18;33(5):795-806. doi: 10.1021/acs.bioconjchem.2c00059. Epub 2022 Apr 21. Bioconjug Chem. 2022. PMID: 35446024 Free PMC article.
-
Metalloproteinases and Their Inhibitors: Potential for the Development of New Therapeutics.Cells. 2020 May 25;9(5):1313. doi: 10.3390/cells9051313. Cells. 2020. PMID: 32466129 Free PMC article. Review.
References
-
- Erijman A, Aizner Y, Shifman JM (2011) Multispecific recognition: mechanism, evolution, and design. Biochemistry 50: 602–611. - PubMed
-
- Brew K, Dinakarpandian D, Nagase H (2000) Tissue inhibitors of metalloproteinases: evolution, structure and function. Biochim Biophys Acta 1477: 267–283. - PubMed
-
- Murphy G, Nagase H (2008) Reappraising metalloproteinases in rheumatoid arthritis and osteoarthritis: destruction or repair? Nat Clin Pract Rheumatol 4: 128–135. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

