Enlightenment of yeast mitochondrial homoplasmy: diversified roles of gene conversion
- PMID: 24710143
- PMCID: PMC3924846
- DOI: 10.3390/genes2010169
Enlightenment of yeast mitochondrial homoplasmy: diversified roles of gene conversion
Abstract
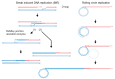
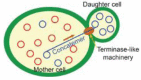
Mitochondria have their own genomic DNA. Unlike the nuclear genome, each cell contains hundreds to thousands of copies of mitochondrial DNA (mtDNA). The copies of mtDNA tend to have heterogeneous sequences, due to the high frequency of mutagenesis, but are quickly homogenized within a cell ("homoplasmy") during vegetative cell growth or through a few sexual generations. Heteroplasmy is strongly associated with mitochondrial diseases, diabetes and aging. Recent studies revealed that the yeast cell has the machinery to homogenize mtDNA, using a common DNA processing pathway with gene conversion; i.e., both genetic events are initiated by a double-stranded break, which is processed into 3' single-stranded tails. One of the tails is base-paired with the complementary sequence of the recipient double-stranded DNA to form a D-loop (homologous pairing), in which repair DNA synthesis is initiated to restore the sequence lost by the breakage. Gene conversion generates sequence diversity, depending on the divergence between the donor and recipient sequences, especially when it occurs among a number of copies of a DNA sequence family with some sequence variations, such as in immunoglobulin diversification in chicken. MtDNA can be regarded as a sequence family, in which the members tend to be diversified by a high frequency of spontaneous mutagenesis. Thus, it would be interesting to determine why and how double-stranded breakage and D-loop formation induce sequence homogenization in mitochondria and sequence diversification in nuclear DNA. We will review the mechanisms and roles of mtDNA homoplasmy, in contrast to nuclear gene conversion, which diversifies gene and genome sequences, to provide clues toward understanding how the common DNA processing pathway results in such divergent outcomes.
Figures


References
-
- Linnane A.W., Marzuki S., Ozawa T., Tanaka M. Mitochondrial DNA mutations as an important contributor to ageing and degenerative diseases. Lancet. 1989;333:642–645. - PubMed
-
- Ozawa T. Genetic and functional changes in mitochondria associated with aging. Physiol. Rev. 1997;77:425–464. - PubMed
-
- Shoubridge E.A., Wai T. Mitochondrial DNA and the mammalian oocyte. Curr. Top. Dev. Biol. 2007;77:87–111. - PubMed
-
- Cao L., Shitara H., Horii T., Nagao Y., Imai H., Abe K., Hara T., Hayashi J.I., Yonekawa H. The mitochondrial bottleneck occurs without reduction of mtDNA content in female mouse germ cells. Nat. Genet. 2007;39:386–390. - PubMed
-
- Cree L.M., Samuels D.C., de Sousa Lopes S.C., Rajasimha H.K., Wonnapinij P., Mann J.R., Dahl H.H., Chinnery P.F. A reduction of mitochondrial DNA molecules during embryogenesis explains the rapid segregation of genotypes. Nat. Genet. 2008;40:249–254. - PubMed
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

