Aberrant Single Exon Skipping is not Altered by Age in Exons of NF1, RABAC1, AATF or PCGF2 in Human Blood Cells and Fibroblasts
- PMID: 24710210
- PMCID: PMC3927615
- DOI: 10.3390/genes2030562
Aberrant Single Exon Skipping is not Altered by Age in Exons of NF1, RABAC1, AATF or PCGF2 in Human Blood Cells and Fibroblasts
Abstract
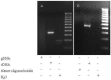
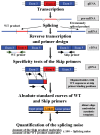
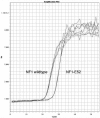
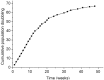
In human pre-mRNA splicing, infrequent errors occur resulting in erroneous splice products as shown in a genome-wide approach. One characteristic subgroup consists of products lacking one cassette exon. The noise in the splicing process, represented by those misspliced products, can be increased by cold shock treatment or by inhibiting the nonsense mediated decay. Here, we investigated whether the splicing noise frequency increases with age in vivo in peripheral bloods cells or in vitro in cultured and aged fibroblasts from healthy donors. Splicing noise frequency was measured for four erroneously skipped NF1 exons and one exon of RABAC1, AATF and PCGF2 by RT-qPCR. Measurements were validated in cultured fibroblasts treated with cold shock or puromycin. Intragenic but not interpersonal differences were detected in splicing noise frequencies in vivo in peripheral blood cells of 11 healthy donors (15 y-85 y) and in in vitro senescent fibroblasts from three further donors. No correlation to the age of the donors was found in the splicing noise frequencies. Our data demonstrates that splicing error frequencies are not altered by age in peripheral blood cells or in vitro aged fibroblasts in the tested exons of the four investigated genes, indicating a high importance of correct splicing in these proliferating aged cells.
Figures
Similar articles
-
Cyclic stretch increases splicing noise rate in cultured human fibroblasts.BMC Res Notes. 2011 Oct 31;4:470. doi: 10.1186/1756-0500-4-470. BMC Res Notes. 2011. PMID: 22040907 Free PMC article.
-
RT-PCR splicing analysis of the NF1 open reading frame.Hum Genet. 2002 May;110(5):495-502. doi: 10.1007/s00439-002-0714-6. Epub 2002 Apr 4. Hum Genet. 2002. PMID: 12073021
-
A novel mutation in the neurofibromatosis type 1 (NF1) gene promotes skipping of two exons by preventing exon definition.J Mol Biol. 2001 Apr 13;307(5):1261-70. doi: 10.1006/jmbi.2001.4561. J Mol Biol. 2001. PMID: 11292340
-
The association of nonsense codons with exon skipping.Mutat Res. 1998 Sep;411(2):87-117. doi: 10.1016/s1383-5742(98)00010-6. Mutat Res. 1998. PMID: 9806422 Review.
-
How prevalent is functional alternative splicing in the human genome?Trends Genet. 2004 Feb;20(2):68-71. doi: 10.1016/j.tig.2003.12.004. Trends Genet. 2004. PMID: 14746986 Review.
Cited by
-
Enhancing endosomal escape of transduced proteins by photochemical internalisation.PLoS One. 2012;7(12):e52473. doi: 10.1371/journal.pone.0052473. Epub 2012 Dec 21. PLoS One. 2012. PMID: 23285056 Free PMC article.
-
Ultra-deep sequencing reveals pre-mRNA splicing as a sequence driven high-fidelity process.PLoS One. 2019 Oct 3;14(10):e0223132. doi: 10.1371/journal.pone.0223132. eCollection 2019. PLoS One. 2019. PMID: 31581208 Free PMC article.
-
Cyclic stretch increases splicing noise rate in cultured human fibroblasts.BMC Res Notes. 2011 Oct 31;4:470. doi: 10.1186/1756-0500-4-470. BMC Res Notes. 2011. PMID: 22040907 Free PMC article.
References
-
- Meshorer E., Soreq H. Pre-mRNA splicing modulations in senescence. Aging Cell. 2002;1:10–16. - PubMed
-
- Hertel K.J. Combinatorial control of exon recognition. J. Biol. Chem. 2008;283:1211–1215. - PubMed
-
- Wimmer K., Eckart M., Rehder H., Fonatsch C. Illegitimate splicing of the NF1 gene in healthy individuals mimics mutation-induced splicing alterations in NF1 patients. Hum. Genet. 2000;106:311–313. - PubMed
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

