Multisite phosphorylation of the Sum1 transcriptional repressor by S-phase kinases controls exit from meiotic prophase in yeast
- PMID: 24710277
- PMCID: PMC4054303
- DOI: 10.1128/MCB.01413-13
Multisite phosphorylation of the Sum1 transcriptional repressor by S-phase kinases controls exit from meiotic prophase in yeast
Abstract
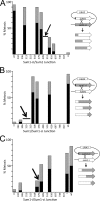
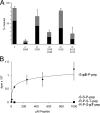
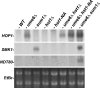
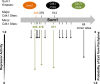
Activation of the meiotic transcription factor Ndt80 is a key regulatory transition in the life cycle of Saccharomyces cerevisiae because it triggers exit from pachytene and entry into meiosis. The NDT80 promoter is held inactive by a complex containing the DNA-binding protein Sum1 and the histone deacetylase Hst1. Meiosis-specific phosphorylation of Sum1 by the protein kinases Cdk1, Ime2, and Cdc7 is required for NDT80 expression. Here, we show that the S-phase-promoting cyclin Clb5 activates Cdk1 to phosphorylate most, and perhaps all, of the 11 minimal cyclin-dependent kinase (CDK) phospho-consensus sites (S/T-P) in Sum1. Nine of these sites can individually promote modest levels of meiosis, yet these sites function in a quasiadditive manner to promote substantial levels of meiosis. Two Cdk1 sites and an Ime2 site individually promote high levels of meiosis, likely by preparing Sum1 for phosphorylation by Cdc7. Chromatin immunoprecipitation reveals that the phosphorylation sites are required for removal of Sum1 from the NDT80 promoter. We also find that Sum1, but not its partner protein Hst1, is required to repress NDT80 transcription. Thus, while the phosphorylation of Sum1 may lead to dissociation from DNA by influencing Hst1, it is the presence of Sum1 on DNA that determines whether NDT80 will be expressed.
Copyright © 2014, American Society for Microbiology. All Rights Reserved.
Figures




References
-
- Nobles KN, Xiao K, Ahn S, Shukla AK, Lam CM, Rajagopal S, Strachan RT, Huang TY, Bressler EA, Hara MR, Shenoy SK, Gygi SP, Lefkowitz RJ. 2011. Distinct phosphorylation sites on the beta(2)-adrenergic receptor establish a barcode that encodes differential functions of beta-arrestin. Sci. Signal. 4:ra51. 10.1126/scisignal.2001707 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous
