The structured core domain of αB-crystallin can prevent amyloid fibrillation and associated toxicity
- PMID: 24711386
- PMCID: PMC4000792
- DOI: 10.1073/pnas.1322673111
The structured core domain of αB-crystallin can prevent amyloid fibrillation and associated toxicity
Abstract
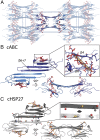
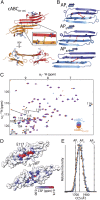
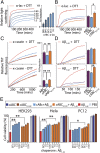
Mammalian small heat-shock proteins (sHSPs) are molecular chaperones that form polydisperse and dynamic complexes with target proteins, serving as a first line of defense in preventing their aggregation into either amorphous deposits or amyloid fibrils. Their apparently broad target specificity makes sHSPs attractive for investigating ways to tackle disorders of protein aggregation. The two most abundant sHSPs in human tissue are αB-crystallin (ABC) and HSP27; here we present high-resolution structures of their core domains (cABC, cHSP27), each in complex with a segment of their respective C-terminal regions. We find that both truncated proteins dimerize, and although this interface is labile in the case of cABC, in cHSP27 the dimer can be cross-linked by an intermonomer disulfide linkage. Using cHSP27 as a template, we have designed an equivalently locked cABC to enable us to investigate the functional role played by oligomerization, disordered N and C termini, subunit exchange, and variable dimer interfaces in ABC. We have assayed the ability of the different forms of ABC to prevent protein aggregation in vitro. Remarkably, we find that cABC has chaperone activity comparable to that of the full-length protein, even when monomer dissociation is restricted through disulfide linkage. Furthermore, cABC is a potent inhibitor of amyloid fibril formation and, by slowing the rate of its aggregation, effectively reduces the toxicity of amyloid-β peptide to cells. Overall we present a small chaperone unit together with its atomic coordinates that potentially enables the rational design of more effective chaperones and amyloid inhibitors.
Keywords: X-ray crystallography; ion mobility mass spectrometry; nuclear magnetic resonance spectroscopy.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


References
-
- Baldwin AJ, et al. Metastability of native proteins and the phenomenon of amyloid formation. J Am Chem Soc. 2011;133(36):14160–14163. - PubMed
-
- Olzscha H, et al. Amyloid-like aggregates sequester numerous metastable proteins with essential cellular functions. Cell. 2011;144(1):67–78. - PubMed
-
- Balch WE, Morimoto RI, Dillin A, Kelly JW. Adapting proteostasis for disease intervention. Science. 2008;319(5865):916–919. - PubMed
-
- Hartl FU, Bracher A, Hayer-Hartl M. Molecular chaperones in protein folding and proteostasis. Nature. 2011;475(7356):324–332. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

