STAT3, STAT4, NFATc1, and CTCF regulate PD-1 through multiple novel regulatory regions in murine T cells
- PMID: 24711622
- PMCID: PMC4011967
- DOI: 10.4049/jimmunol.1302750
STAT3, STAT4, NFATc1, and CTCF regulate PD-1 through multiple novel regulatory regions in murine T cells
Abstract
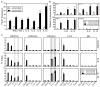
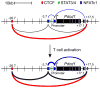
Programmed death-1 (PD-1) is a crucial negative regulator of CD8 T cell development and function, yet the mechanisms that control its expression are not fully understood. Through a nonbiased DNase I hypersensitivity assay, four novel regulatory regions within the Pdcd1 locus were identified. Two of these elements flanked the locus, bound the transcriptional insulator protein CCCTC-binding factor, and interacted with each other, creating a potential regulatory compartmentalization of the locus. In response to T cell activation signaling, NFATc1 bound to two of the novel regions that function as independent regulatory elements. STAT binding sites were identified in these elements as well. In splenic CD8 T cells, TCR-induced PD-1 expression was augmented by IL-6 and IL-12, inducers of STAT3 and STAT4 activity, respectively. IL-6 or IL-12 on its own did not induce PD-1. Importantly, STAT3/4 and distinct chromatin modifications were associated with the novel regulatory regions following cytokine stimulation. The NFATc1/STAT regulatory regions were found to interact with the promoter region of the Pdcd1 gene, providing a mechanism for their action. Together these data add multiple novel distal regulatory regions and pathways to the control of PD-1 expression and provide a molecular mechanism by which proinflammatory cytokines, such as IL-6 or IL-12, can augment PD-1 expression.
Figures





References
-
- Day CL, Kaufmann DE, Kiepiela P, Brown JA, Moodley ES, Reddy S, Mackey EW, Miller JD, Leslie AJ, DePierres C, Mncube Z, Duraiswamy J, Zhu B, Eichbaum Q, Altfeld M, Wherry EJ, Coovadia HM, Goulder PJR, Klenerman P, Ahmed R, Freeman GJ, Walker BD. PD-1 expression on HIV-specific T cells is associated with T-cell exhaustion and disease progression. Nature. 2006;443:350–354. - PubMed
-
- Fourcade J, Sun Z, Benallaoua M, Guillaume P, Luescher IF, Sander C, Kirkwood JM, Kuchroo V, Zarour HM. Upregulation of Tim-3 and PD-1 expression is associated with tumor antigen‚Äìspecific CD8+ T cell dysfunction in melanoma patients. The Journal of Experimental Medicine. 2010;207:2175–2186. - PMC - PubMed
-
- Wherry EJ, Ha SJ, Kaech SM, Haining WN, Sarkar S, Kalia V, Subramaniam S, Blattman JN, Barber DL, Ahmed R. Molecular Signature of CD8+ T Cell Exhaustion during Chronic Viral Infection. Immunity. 2007;27:670–684. - PubMed
-
- Akbay EA, Koyama S, Carretero J, Altabef A, Tchaicha JH, Christensen CL, Mikse OR, Cherniack AD, Beauchamp EM, Pugh TJ, Wilkerson MD, Fecci PE, Butaney M, Reibel JB, Soucheray M, Cohoon TJ, Janne PA, Meyerson M, Hayes DN, Shapiro GI, Shimamura T, Sholl LM, Rodig SJ, Freeman GJ, Hammerman PS, Dranoff G, Wong KK. Activation of the PD-1 pathway contributes to immune escape in EGFR-driven lung tumors. Cancer discovery 2013 - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

