Next-generation in situ hybridization chain reaction: higher gain, lower cost, greater durability
- PMID: 24712299
- PMCID: PMC4046802
- DOI: 10.1021/nn405717p
Next-generation in situ hybridization chain reaction: higher gain, lower cost, greater durability
Abstract
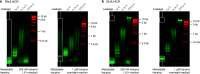
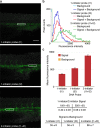
Hybridization chain reaction (HCR) provides multiplexed, isothermal, enzyme-free, molecular signal amplification in diverse settings. Within intact vertebrate embryos, where signal-to-background is at a premium, HCR in situ amplification enables simultaneous mapping of multiple target mRNAs, addressing a longstanding challenge in the biological sciences. With this approach, RNA probes complementary to mRNA targets trigger chain reactions in which metastable fluorophore-labeled RNA hairpins self-assemble into tethered fluorescent amplification polymers. The properties of HCR lead to straightforward multiplexing, deep sample penetration, high signal-to-background, and sharp subcellular signal localization within fixed whole-mount zebrafish embryos, a standard model system for the study of vertebrate development. However, RNA reagents are expensive and vulnerable to enzymatic degradation. Moreover, the stringent hybridization conditions used to destabilize nonspecific hairpin binding also reduce the energetic driving force for HCR polymerization, creating a trade-off between minimization of background and maximization of signal. Here, we eliminate this trade-off by demonstrating that low background levels can be achieved using permissive in situ amplification conditions (0% formamide, room temperature) and engineer next-generation DNA HCR amplifiers that maximize the free energy benefit per polymerization step while preserving the kinetic trapping property that underlies conditional polymerization, dramatically increasing signal gain, reducing reagent cost, and improving reagent durability.
Figures






References
-
- Qian X.; Jin L.; Lloyd R. V. In Situ Hybridization: Basic Approaches and Recent Development. J. Histotechnol. 2004, 27, 53–67.
-
- Silverman A.; Kool E. Oligonucleotide Probes for RNA-Targeted Fluorescence in Situ Hybridization. Adv. Clin. Chem. 2007, 43, 79–115. - PubMed
-
- Femino A.; Fay F. S.; Fogarty K.; Singer R. H. Visualization of Single RNA Transcripts in Situ. Science 1998, 280, 585–590. - PubMed
-
- Levsky J. M.; Shenoy S. M.; Pezo R. C.; Singer R. H. Single-Cell Gene Expression Profiling. Science 2002, 297, 836–840. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

