Pulsatile exposure to simulated reflux leads to changes in gene expression in a 3D model of oesophageal mucosa
- PMID: 24713057
- PMCID: PMC4351858
- DOI: 10.1111/iep.12083
Pulsatile exposure to simulated reflux leads to changes in gene expression in a 3D model of oesophageal mucosa
Abstract
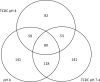
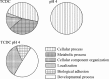
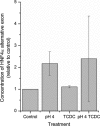
Oesophageal exposure to duodenogastroesophageal refluxate is implicated in the development of Barrett's metaplasia (BM), with increased risk of progression to oesophageal adenocarcinoma. The literature proposes that reflux exposure activates NF-κB, driving the aberrant expression of intestine-specific caudal-related homeobox (CDX) genes. However, early events in the pathogenesis of BM from normal epithelium are poorly understood. To investigate this, our study subjected a 3D model of the normal human oesophageal mucosa to repeated, pulsatile exposure to specific bile components and examined changes in gene expression. Initial 2D experiments with a range of bile salts observed that taurochenodeoxycholate (TCDC) impacted upon NF-κB activation without causing cell death. Informed by this, the 3D oesophageal model was repeatedly exposed to TCDC in the presence and absence of acid, and the epithelial cells underwent gene expression profiling. We identified ~300 differentially expressed genes following each treatment, with a large and significant overlap between treatments. Enrichment analysis (Broad GSEA, DAVID and Metacore™; GeneGo Inc) identified multiple gene sets related to cell signalling, inflammation, proliferation, differentiation and cell adhesion. Specifically NF-κB activation, Wnt signalling, cell adhesion and targets for the transcription factors PTF1A and HNF4α were highlighted. Our data suggest that HNF4α isoform switching may be an early event in Barrett's pathogenesis. CDX1/2 targets were, however, not enriched, suggesting that although CDX1/2 activation reportedly plays a role in BM development, it may not be an initial event. Our findings highlight new areas for investigation in the earliest stages of BM pathogenesis of oesophageal diseases and new potential therapeutic targets.
Keywords: Barrett's metaplasia; HNF4alpha; acid; oesophageal adenocarcinoma; reflux; taurochenodeoxycholate; tissue engineering.
© 2014 The Authors. International Journal of Experimental Pathology © 2014 International Journal of Experimental Pathology.
Figures




Similar articles
-
Aberrant epithelial-mesenchymal Hedgehog signaling characterizes Barrett's metaplasia.Gastroenterology. 2010 May;138(5):1810-22. doi: 10.1053/j.gastro.2010.01.048. Epub 2010 Feb 4. Gastroenterology. 2010. PMID: 20138038 Free PMC article.
-
Bile acid-induced expression of activation-induced cytidine deaminase during the development of Barrett's oesophageal adenocarcinoma.Carcinogenesis. 2011 Nov;32(11):1706-12. doi: 10.1093/carcin/bgr194. Epub 2011 Sep 1. Carcinogenesis. 2011. PMID: 21890457
-
Roles of caudal-related homeobox gene Cdx1 in oesophageal epithelial cells in Barrett's epithelium development.Gut. 2009 May;58(5):620-8. doi: 10.1136/gut.2008.152975. Epub 2009 Jan 9. Gut. 2009. PMID: 19136512
-
Barrett's oesophagus and oesophageal adenocarcinoma: how does acid interfere with cell proliferation and differentiation?Gut. 2005 Mar;54 Suppl 1(Suppl 1):i21-6. doi: 10.1136/gut.2004.041558. Gut. 2005. PMID: 15711004 Free PMC article. Review.
-
Reflux esophagitis and its role in the pathogenesis of Barrett's metaplasia.J Gastroenterol. 2017 Jul;52(7):767-776. doi: 10.1007/s00535-017-1342-1. Epub 2017 Apr 27. J Gastroenterol. 2017. PMID: 28451845 Free PMC article. Review.
Cited by
-
Rocking media over ex vivo corneas improves this model and allows the study of the effect of proinflammatory cytokines on wound healing.Invest Ophthalmol Vis Sci. 2015 Feb 5;56(3):1553-61. doi: 10.1167/iovs.14-15308. Invest Ophthalmol Vis Sci. 2015. PMID: 25655804 Free PMC article.
-
TGR5-HNF4α axis contributes to bile acid-induced gastric intestinal metaplasia markers expression.Cell Death Discov. 2020 Jul 6;6:56. doi: 10.1038/s41420-020-0290-3. eCollection 2020. Cell Death Discov. 2020. PMID: 32655894 Free PMC article.
-
Production, Characterization and Potential Uses of a 3D Tissue-engineered Human Esophageal Mucosal Model.J Vis Exp. 2015 May 18;(99):e52693. doi: 10.3791/52693. J Vis Exp. 2015. PMID: 26067284 Free PMC article.
-
Controlled bile acid exposure to oesophageal mucosa causes up-regulation of nuclear γ-H2AX possibly via iNOS induction.Biosci Rep. 2016 Jul 7;36(4):e00357. doi: 10.1042/BSR20160124. Print 2016 Aug. Biosci Rep. 2016. PMID: 27247425 Free PMC article.
-
Hnf4α is a key gene that can generate columnar metaplasia in oesophageal epithelium.Differentiation. 2017 Jan-Feb;93:39-49. doi: 10.1016/j.diff.2016.11.001. Epub 2016 Nov 19. Differentiation. 2017. PMID: 27875772 Free PMC article.
References
-
- Babeu JP, Darsigny M, Lussier CR, Boudreau F. Hepatocyte nuclear factor 4alpha contributes to an intestinal epithelial phenotype in vitro and plays a partial role in mouse intestinal epithelium differentiation. Am. J. Physiol. Gastrointest. Liver Physiol. 2009;297:G124–G134. - PubMed
-
- Benahmed F, Gross I, Gaunt SJ, et al. Multiple regulatory regions control the complex expression pattern of the mouse Cdx2 homeobox gene. Gastroenterology. 2008;135:1238–1247. 1247. - PubMed
-
- Chen KH, Mukaisho K, Sugihara H, Araki Y, Yamamoto G, Hattori T. High animal-fat intake changes the bile-acid composition of bile juice and enhances the development of Barrett's esophagus and esophageal adenocarcinoma in a rat duodenal-contents reflux model. Cancer Sci. 2007;98:1683–1688. - PMC - PubMed
-
- Colleypriest BJ, Farrant JM, Slack JM, Tosh D. Hepatocyte Nuclear Factor 4a (HNF4a) provokes intestinal genes in squamous oesophageal cells. Gut. 2011;60:A171–A172.
-
- Debruyne PR, Witek M, Gong L, et al. Bile acids induce ectopic expression of intestinal guanylyl cyclase C Through nuclear factor-kappaB and Cdx2 in human esophageal cells. Gastroenterology. 2006;130:1191–1206. - PubMed
Publication types
MeSH terms
Substances
Supplementary concepts
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials

