Synergistic and antagonistic drug combinations depend on network topology
- PMID: 24713621
- PMCID: PMC3979733
- DOI: 10.1371/journal.pone.0093960
Synergistic and antagonistic drug combinations depend on network topology
Abstract
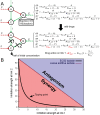
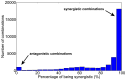
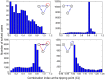
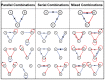
Drug combinations may exhibit synergistic or antagonistic effects. Rational design of synergistic drug combinations remains a challenge despite active experimental and computational efforts. Because drugs manifest their action via their targets, the effects of drug combinations should depend on the interaction of their targets in a network manner. We therefore modeled the effects of drug combinations along with their targets interacting in a network, trying to elucidate the relationships between the network topology involving drug targets and drug combination effects. We used three-node enzymatic networks with various topologies and parameters to study two-drug combinations. These networks can be simplifications of more complex networks involving drug targets, or closely connected target networks themselves. We found that the effects of most of the combinations were not sensitive to parameter variation, indicating that drug combinational effects largely depend on network topology. We then identified and analyzed consistent synergistic or antagonistic drug combination motifs. Synergistic motifs encompass a diverse range of patterns, including both serial and parallel combinations, while antagonistic combinations are relatively less common and homogenous, mostly composed of a positive feedback loop and a downstream link. Overall our study indicated that designing novel synergistic drug combinations based on network topology could be promising, and the motifs we identified could be a useful catalog for rational drug combination design in enzymatic systems.
Conflict of interest statement
Figures
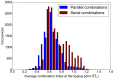

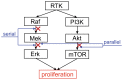
Similar articles
-
Computational analysis of synergism in small networks with different logic.J Biol Phys. 2023 Mar;49(1):1-27. doi: 10.1007/s10867-022-09620-0. Epub 2022 Dec 29. J Biol Phys. 2023. PMID: 36580168 Free PMC article.
-
Biochemical reaction network topology defines dose-dependent Drug-Drug interactions.Comput Biol Med. 2023 Mar;155:106584. doi: 10.1016/j.compbiomed.2023.106584. Epub 2023 Jan 28. Comput Biol Med. 2023. PMID: 36805215
-
Neighbor communities in drug combination networks characterize synergistic effect.Mol Biosyst. 2012 Oct 30;8(12):3185-96. doi: 10.1039/c2mb25267h. Mol Biosyst. 2012. PMID: 23014807
-
Biomolecular Network-Based Synergistic Drug Combination Discovery.Biomed Res Int. 2016;2016:8518945. doi: 10.1155/2016/8518945. Epub 2016 Nov 7. Biomed Res Int. 2016. PMID: 27891522 Free PMC article. Review.
-
Advances in computational approaches in identifying synergistic drug combinations.Brief Bioinform. 2018 Nov 27;19(6):1172-1182. doi: 10.1093/bib/bbx047. Brief Bioinform. 2018. PMID: 28475767 Review.
Cited by
-
The MEK1/2 Inhibitor ATR-002 (Zapnometinib) Synergistically Potentiates the Antiviral Effect of Direct-Acting Anti-SARS-CoV-2 Drugs.Pharmaceutics. 2022 Aug 25;14(9):1776. doi: 10.3390/pharmaceutics14091776. Pharmaceutics. 2022. PMID: 36145524 Free PMC article.
-
Mechanistic model of MAPK signaling reveals how allostery and rewiring contribute to drug resistance.Mol Syst Biol. 2023 Feb 10;19(2):e10988. doi: 10.15252/msb.202210988. Epub 2023 Jan 26. Mol Syst Biol. 2023. PMID: 36700386 Free PMC article.
-
Interpreting the Mechanism of Synergism for Drug Combinations Using Attention-Based Hierarchical Graph Pooling.Cancers (Basel). 2023 Aug 22;15(17):4210. doi: 10.3390/cancers15174210. Cancers (Basel). 2023. PMID: 37686486 Free PMC article.
-
Leaf ethanolic extract of Etlingera hemesphaerica Blume alters mercuric chloride teratogenicity during the post-implantation period in Mus musculus.Toxicol Res. 2019 Nov 21;36(2):131-138. doi: 10.1007/s43188-019-00010-8. eCollection 2020 Apr. Toxicol Res. 2019. PMID: 32257925 Free PMC article.
-
Automated and miniaturized screening of antibiotic combinations via robotic-printed combinatorial droplet platform.Acta Pharm Sin B. 2024 Apr;14(4):1801-1813. doi: 10.1016/j.apsb.2023.11.027. Epub 2023 Nov 28. Acta Pharm Sin B. 2024. PMID: 38572105 Free PMC article.
References
-
- Feala JD, Cortes J, Duxbury PM, Piermarocchi C, McCulloch AD, et al. (2010) Systems approaches and algorithms for discovery of combinatorial therapies. Wiley Interdiscip Rev Syst Biol Med 2: 181–193. - PubMed
-
- Fitzgerald JB, Schoeberl B, Nielsen UB, Sorger PK (2006) Systems biology and combination therapy in the quest for clinical efficacy. Nat Chem Biol 2: 458–466. - PubMed
-
- Keith CT, Borisy AA, Stockwell BR (2005) Multicomponent therapeutics for networked systems. Nat Rev Drug Discov 4: 71–78. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

