Autophagy-regulating TP53INP2 mediates muscle wasting and is repressed in diabetes
- PMID: 24713655
- PMCID: PMC4001546
- DOI: 10.1172/JCI72327
Autophagy-regulating TP53INP2 mediates muscle wasting and is repressed in diabetes
Abstract
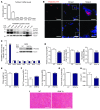
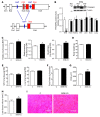
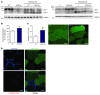
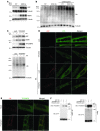
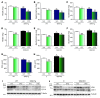
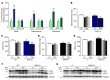
A precise balance between protein degradation and synthesis is essential to preserve skeletal muscle mass. Here, we found that TP53INP2, a homolog of the Drosophila melanogaster DOR protein that regulates autophagy in cellular models, has a direct impact on skeletal muscle mass in vivo. Using different transgenic mouse models, we demonstrated that muscle-specific overexpression of Tp53inp2 reduced muscle mass, while deletion of Tp53inp2 resulted in muscle hypertrophy. TP53INP2 activated basal autophagy in skeletal muscle and sustained p62-independent autophagic degradation of ubiquitinated proteins. Animals with muscle-specific overexpression of Tp53inp2 exhibited enhanced muscle wasting in streptozotocin-induced diabetes that was dependent on autophagy; however, TP53INP2 ablation mitigated experimental diabetes-associated muscle loss. The overexpression or absence of TP53INP2 did not affect muscle wasting in response to denervation, a condition in which autophagy is blocked, further indicating that TP53INP2 alters muscle mass by activating autophagy. Moreover, TP53INP2 expression was markedly repressed in muscle from patients with type 2 diabetes and in murine models of diabetes. Our results indicate that TP53INP2 negatively regulates skeletal muscle mass through activation of autophagy. Furthermore, we propose that TP53INP2 repression is part of an adaptive mechanism aimed at preserving muscle mass under conditions in which insulin action is deficient.
Figures

References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

