Staphylococcal SplB serine protease utilizes a novel molecular mechanism of activation
- PMID: 24713703
- PMCID: PMC4140910
- DOI: 10.1074/jbc.M113.507616
Staphylococcal SplB serine protease utilizes a novel molecular mechanism of activation
Abstract
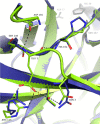
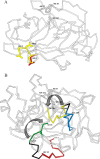
Staphylococcal SplB protease belongs to the chymotrypsin family. Chymotrypsin zymogen is activated by proteolytic processing at the N terminus, resulting in significant structural rearrangement at the active site. Here, we demonstrate that the molecular mechanism of SplB protease activation differs significantly and we characterize the novel mechanism in detail. Using peptide and protein substrates we show that the native signal peptide, or any N-terminal extension, has an inhibitory effect on SplB. Only precise N-terminal processing releases the full proteolytic activity of the wild type analogously to chymotrypsin. However, comparison of the crystal structures of mature SplB and a zymogen mimic show no rearrangement at the active site whatsoever. Instead, only the formation of a unique hydrogen bond network, distant form the active site, by the new N-terminal glutamic acid of mature SplB is observed. The importance of this network and influence of particular hydrogen bond interactions at the N terminus on the catalytic process is demonstrated by evaluating the kinetics of a series of mutants. The results allow us to propose a consistent model where changes in the overall protein dynamics rather than structural rearrangement of the active site are involved in the activation process.
Keywords: Crystal Structure; Enzyme Inactivation; Protease; Serine Protease; Signal Peptide; Staphylococcus aureus; Zymogen Activation.
© 2014 by The American Society for Biochemistry and Molecular Biology, Inc.
Figures

References
-
- Strausberg S., Alexander P., Wang L., Schwarz F., Bryan P. (1993) Catalysis of a protein folding reaction: thermodynamic and kinetic analysis of subtilisin BPN′ interactions with its propeptide fragment. Biochemistry 32, 8112–8119 - PubMed
-
- Baker D., Shiau A. K., Agard D. A. (1993) The role of pro regions in protein folding. Curr. Op. Cell Biol. 5, 966–970 - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources

