Susceptibility to chronic mucus hypersecretion, a genome wide association study
- PMID: 24714607
- PMCID: PMC3979657
- DOI: 10.1371/journal.pone.0091621
Susceptibility to chronic mucus hypersecretion, a genome wide association study
Erratum in
-
Correction: Susceptibility to chronic mucus hypersecretion, a genome wide association study.PLoS One. 2015 May 29;10(5):e0129524. doi: 10.1371/journal.pone.0129524. eCollection 2015. PLoS One. 2015. PMID: 26024482 Free PMC article. No abstract available.
Abstract
Background: Chronic mucus hypersecretion (CMH) is associated with an increased frequency of respiratory infections, excess lung function decline, and increased hospitalisation and mortality rates in the general population. It is associated with smoking, but it is unknown why only a minority of smokers develops CMH. A plausible explanation for this phenomenon is a predisposing genetic constitution. Therefore, we performed a genome wide association (GWA) study of CMH in Caucasian populations.
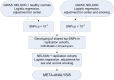
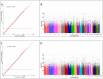
Methods: GWA analysis was performed in the NELSON-study using the Illumina 610 array, followed by replication and meta-analysis in 11 additional cohorts. In total 2,704 subjects with, and 7,624 subjects without CMH were included, all current or former heavy smokers (≥20 pack-years). Additional studies were performed to test the functional relevance of the most significant single nucleotide polymorphism (SNP).
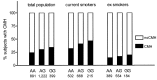
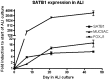
Results: A strong association with CMH, consistent across all cohorts, was observed with rs6577641 (p = 4.25×10(-6), OR = 1.17), located in intron 9 of the special AT-rich sequence-binding protein 1 locus (SATB1) on chromosome 3. The risk allele (G) was associated with higher mRNA expression of SATB1 (4.3×10(-9)) in lung tissue. Presence of CMH was associated with increased SATB1 mRNA expression in bronchial biopsies from COPD patients. SATB1 expression was induced during differentiation of primary human bronchial epithelial cells in culture.
Conclusions: Our findings, that SNP rs6577641 is associated with CMH in multiple cohorts and is a cis-eQTL for SATB1, together with our additional observation that SATB1 expression increases during epithelial differentiation provide suggestive evidence that SATB1 is a gene that affects CMH.
Conflict of interest statement
Figures


References
-
- Anonymous (1995) Standards for the diagnosis and care of patients with chronic obstructive pulmonary disease. american thoracic society. Am J Respir Crit Care Med 152: S77–121. - PubMed
-
- Ferre A, Fuhrman C, Zureik M, Chouaid C, Vergnenegre A, et al. (2012) Chronic bronchitis in the general population: Influence of age, gender and socio-economic conditions. Respir Med 106: 467–471. - PubMed
-
- Rabe KF, Hurd S, Anzueto A, Barnes PJ, Buist SA, et al. (2007) Global strategy for the diagnosis, management, and prevention of chronic obstructive pulmonary disease: GOLD executive summary. Am J Respir Crit Care Med 176: 532–555. - PubMed
-
- Vestbo J, Rasmussen FV (1989) Respiratory symptoms and FEV1 as predictors of hospitalization and medication in the following 12 years due to respiratory disease. Eur Respir J 2: 710–715. - PubMed
-
- Vestbo J, Prescott E, Lange P (1996) Association of chronic mucus hypersecretion with FEV1 decline and chronic obstructive pulmonary disease morbidity. copenhagen city heart study group. Am J Respir Crit Care Med 153: 1530–1535. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials

