Gene activation and cell fate control in plants: a chromatin perspective
- PMID: 24714879
- PMCID: PMC11113918
- DOI: 10.1007/s00018-014-1609-0
Gene activation and cell fate control in plants: a chromatin perspective
Abstract
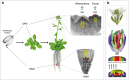
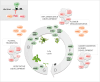
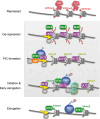
In plants, environment-adaptable organogenesis extends throughout the lifespan, and iterative development requires repetitive rounds of activation and repression of several sets of genes. Eukaryotic genome compaction into chromatin forms a physical barrier for transcription; therefore, induction of gene expression requires alteration in chromatin structure. One of the present great challenges in molecular and developmental biology is to understand how chromatin is brought from a repressive to permissive state on specific loci and in a very specific cluster of cells, as well as how this state is further maintained and propagated through time and cell division in a cell lineage. In this review, we report recent discoveries implementing our knowledge on chromatin dynamics that modulate developmental gene expression. We also discuss how new data sets highlight plant specificities, likely reflecting requirement for a highly dynamic chromatin.
Figures
References
-
- Fornara F., Montaigu A., Coupland G. SnapShot: Control of flowering in Arabidopsis. Cell. 2010;141:550. - PubMed
-
- Kaufmann K, Nagasaki M, Jáuregui R. Modelling the molecular interactions in the flower developmental network of Arabidopsis thaliana. In Silico Biol. 2010;10:125–143. - PubMed
-
- Posé D, Yant L, Schmid M. The end of innocence: flowering networks explode in complexity. Curr Opin Plant Biol. 2012;15:45–50. - PubMed
-
- Coen ES, Meyerowitz EM. The war of the whorls: genetic interactions controlling flower development. Nature. 1991;353:31–37. - PubMed
-
- Krizek BA, Fletcher JC. Molecular mechanisms of flower development: an armchair guide. Nat Rev Genet. 2005;6:688–698. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

