Outer membrane β-barrel protein folding is physically controlled by periplasmic lipid head groups and BamA
- PMID: 24715731
- PMCID: PMC4000854
- DOI: 10.1073/pnas.1322473111
Outer membrane β-barrel protein folding is physically controlled by periplasmic lipid head groups and BamA
Abstract
Outer membrane β-barrel proteins (OMPs) are crucial for numerous cellular processes in prokaryotes and eukaryotes. Despite extensive studies on OMP biogenesis, it is unclear why OMPs require assembly machineries to fold into their native outer membranes, as they are capable of folding quickly and efficiently through an intrinsic folding pathway in vitro. By investigating the folding of several bacterial OMPs using membranes with naturally occurring Escherichia coli lipids, we show that phosphoethanolamine and phosphoglycerol head groups impose a kinetic barrier to OMP folding. The kinetic retardation of OMP folding places a strong negative pressure against spontaneous incorporation of OMPs into inner bacterial membranes, which would dissipate the proton motive force and undoubtedly kill bacteria. We further show that prefolded β-barrel assembly machinery subunit A (BamA), the evolutionarily conserved, central subunit of the BAM complex, accelerates OMP folding by lowering the kinetic barrier imposed by phosphoethanolamine head groups. Our results suggest that OMP assembly machineries are required in vivo to enable physical control over the spontaneously occurring OMP folding reaction in the periplasm. Mechanistic studies further allowed us to derive a model for BamA function, which explains how OMP assembly can be conserved between prokaryotes and eukaryotes.
Keywords: beta-barrel transmembrane protein; membrane protein folding.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
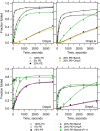

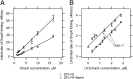


References
-
- Tamm LK, Hong H, Liang B. Folding and assembly of beta-barrel membrane proteins. Biochim Biophys Acta. 2004;1666(1-2):250–263. - PubMed
-
- Wimley WC. The versatile beta-barrel membrane protein. Curr Opin Struct Biol. 2003;13(4):404–411. - PubMed
-
- Bajaj H, et al. Antibiotic uptake through membrane channels: Role of Providencia stuartii OmpPst1 porin in carbapenem resistance. Biochemistry. 2012;51(51):10244–10249. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

