Palpitomonas bilix represents a basal cryptist lineage: insight into the character evolution in Cryptista
- PMID: 24717814
- PMCID: PMC3982174
- DOI: 10.1038/srep04641
Palpitomonas bilix represents a basal cryptist lineage: insight into the character evolution in Cryptista
Abstract
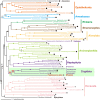
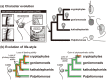
Phylogenetic position of the marine biflagellate Palpitomonas bilix is intriguing, since several ultrastructural characteristics implied its evolutionary connection to Archaeplastida or Hacrobia. The origin and early evolution of these two eukaryotic assemblages have yet to be fully elucidated, and P. bilix may be a key lineage in tracing those groups' early evolution. In the present study, we analyzed a 'phylogenomic' alignment of 157 genes to clarify the position of P. bilix in eukaryotic phylogeny. In the 157-gene phylogeny, P. bilix was found to be basal to a clade of cryptophytes, goniomonads and kathablepharids, collectively known as Cryptista, which is proposed to be a part of the larger taxonomic assemblage Hacrobia. We here discuss the taxonomic assignment of P. bilix, and character evolution in Cryptista.
Figures

Similar articles
-
Palpitomonas bilix gen. et sp. nov.: A novel deep-branching heterotroph possibly related to Archaeplastida or Hacrobia.Protist. 2010 Oct;161(4):523-38. doi: 10.1016/j.protis.2010.03.001. Epub 2010 Apr 24. Protist. 2010. PMID: 20418156
-
Mitochondrial Genome of Palpitomonas bilix: Derived Genome Structure and Ancestral System for Cytochrome c Maturation.Genome Biol Evol. 2016 Oct 13;8(10):3090-3098. doi: 10.1093/gbe/evw217. Genome Biol Evol. 2016. PMID: 27604877 Free PMC article.
-
Molecular phylogeny and description of the novel katablepharid Roombia truncata gen. et sp. nov., and establishment of the Hacrobia taxon nov.PLoS One. 2009 Sep 17;4(9):e7080. doi: 10.1371/journal.pone.0007080. PLoS One. 2009. PMID: 19759916 Free PMC article.
-
Nucleomorph genomes.Annu Rev Genet. 2009;43:251-64. doi: 10.1146/annurev-genet-102108-134809. Annu Rev Genet. 2009. PMID: 19686079 Review.
-
Phytochrome evolution in 3D: deletion, duplication, and diversification.New Phytol. 2020 Mar;225(6):2283-2300. doi: 10.1111/nph.16240. Epub 2019 Nov 2. New Phytol. 2020. PMID: 31595505 Free PMC article. Review.
Cited by
-
Updating algal evolutionary relationships through plastid genome sequencing: did alveolate plastids emerge through endosymbiosis of an ochrophyte?Sci Rep. 2015 May 28;5:10134. doi: 10.1038/srep10134. Sci Rep. 2015. PMID: 26017773 Free PMC article.
-
Sex is a ubiquitous, ancient, and inherent attribute of eukaryotic life.Proc Natl Acad Sci U S A. 2015 Jul 21;112(29):8827-34. doi: 10.1073/pnas.1501725112. Proc Natl Acad Sci U S A. 2015. PMID: 26195746 Free PMC article.
-
Comparative Plastid Genomics of Cryptomonas Species Reveals Fine-Scale Genomic Responses to Loss of Photosynthesis.Genome Biol Evol. 2020 Feb 1;12(2):3926-3937. doi: 10.1093/gbe/evaa001. Genome Biol Evol. 2020. PMID: 31922581 Free PMC article.
-
Comparative mitochondrial genomics of cryptophyte algae: gene shuffling and dynamic mobile genetic elements.BMC Genomics. 2018 Apr 20;19(1):275. doi: 10.1186/s12864-018-4626-9. BMC Genomics. 2018. PMID: 29678149 Free PMC article.
-
Evolutionary trajectory for nuclear functions of ciliary transport complex proteins.Microbiol Mol Biol Rev. 2024 Sep 26;88(3):e0000624. doi: 10.1128/mmbr.00006-24. Epub 2024 Jul 12. Microbiol Mol Biol Rev. 2024. PMID: 38995044 Free PMC article. Review.
References
-
- Moore R. B. et al. A photosynthetic alveolate closely related to apicomplexan parasites. Nature. 451, 959–963 (2008). - PubMed
-
- Yabuki A., Ishida K. & Cavalier-Smith T. Rigifila ramosa n. gen., n. sp. Filose amoeba with a distinct pellicle, is related to Micronuclearia. Protist. 164, 75–88 (2013). - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

