Cullin4A and cullin4B are interchangeable for HIV Vpr and Vpx action through the CRL4 ubiquitin ligase complex
- PMID: 24719410
- PMCID: PMC4054339
- DOI: 10.1128/JVI.00241-14
Cullin4A and cullin4B are interchangeable for HIV Vpr and Vpx action through the CRL4 ubiquitin ligase complex
Abstract
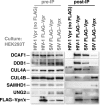
Human immunodeficiency virus (HIV) seizes control of cellular cullin-RING E3 ubiquitin ligases (CRLs) to promote viral replication. HIV-1 Vpr and HIV-2/simian immunodeficiency virus (SIV) Vpr and Vpx engage the cullin4 (CUL4)-containing ubiquitin ligase complex (CRL4) to cause polyubiquitination and proteasomal degradation of host proteins, including ones that block infection. HIV-1 Vpr engages CRL4 to trigger the degradation of uracil-N-glycosylase 2 (UNG2). Both HIV-1 Vpr and HIV-2/SIV Vpr tap CRL4 to initiate G2 cell cycle arrest. HIV-2/SIV Vpx secures CRL4 to degrade the antiviral protein SAMHD1. CRL4 includes either cullin4A (CUL4A) or cullin4B (CUL4B) among its components. Whether Vpr or Vpx relies on CUL4A, CUL4B, or both to act through CRL4 is not known. Reported structural, phenotypic, and intracellular distribution differences between the two CUL4 types led us to hypothesize that Vpr and Vpx employ these in a function-specific manner. Here we determined CUL4 requirements for HIV-1 and HIV-2/SIV Vpr-mediated G2 cell cycle arrest, HIV-1 Vpr-mediated UNG2 degradation, and HIV-2 Vpx-mediated SAMHD1 degradation. Surprisingly, CUL4A and CUL4B are exchangeable for CRL4-dependent Vpr and Vpx action, except in primary macrophages, where Vpx relies on both CUL4A and CUL4B for maximal SAMHD1 depletion. This work highlights the need to consider both CUL4 types for Vpr and Vpx functions and also shows that the intracellular distribution of CUL4A and CUL4B can vary by cell type.
Importance: The work presented here shows for the first time that HIV Vpr and Vpx do not rely exclusively on CUL4A to cause ubiquitination through the CRL4 ubiquitin ligase complex. Furthermore, our finding that intracellular CUL4 and SAMHD1 distributions can vary with cell type provides the basis for reconciling previous disparate findings regarding the site of SAMHD1 depletion. Finally, our observations with primary immune cells provide insight into the cell biology of CUL4A and CUL4B that will help differentiate the functions of these similar proteins.
Copyright © 2014, American Society for Microbiology. All Rights Reserved.
Figures






Similar articles
-
CRL4-DCAF1 Ubiquitin Ligase Dependent Functions of HIV Viral Protein R and Viral Protein X.Viruses. 2024 Aug 17;16(8):1313. doi: 10.3390/v16081313. Viruses. 2024. PMID: 39205287 Free PMC article. Review.
-
Inhibition of Vpx-Mediated SAMHD1 and Vpr-Mediated Host Helicase Transcription Factor Degradation by Selective Disruption of Viral CRL4 (DCAF1) E3 Ubiquitin Ligase Assembly.J Virol. 2017 Apr 13;91(9):e00225-17. doi: 10.1128/JVI.00225-17. Print 2017 May 1. J Virol. 2017. PMID: 28202763 Free PMC article.
-
HIV-1 Vpr loads uracil DNA glycosylase-2 onto DCAF1, a substrate recognition subunit of a cullin 4A-ring E3 ubiquitin ligase for proteasome-dependent degradation.J Biol Chem. 2010 Nov 26;285(48):37333-41. doi: 10.1074/jbc.M110.133181. Epub 2010 Sep 24. J Biol Chem. 2010. PMID: 20870715 Free PMC article.
-
A novel DCAF1-binding motif required for Vpx-mediated degradation of nuclear SAMHD1 and Vpr-induced G2 arrest.Cell Microbiol. 2012 Nov;14(11):1745-56. doi: 10.1111/j.1462-5822.2012.01835.x. Epub 2012 Aug 9. Cell Microbiol. 2012. PMID: 22776683
-
Lentivirus Vpr and Vpx accessory proteins usurp the cullin4-DDB1 (DCAF1) E3 ubiquitin ligase.Curr Opin Virol. 2012 Dec;2(6):755-63. doi: 10.1016/j.coviro.2012.09.010. Epub 2012 Oct 10. Curr Opin Virol. 2012. PMID: 23062609 Free PMC article. Review.
Cited by
-
CUL4B promotes the pathology of adjuvant-induced arthritis in rats through the canonical Wnt signaling.J Mol Med (Berl). 2018 Jun;96(6):495-511. doi: 10.1007/s00109-018-1635-8. Epub 2018 Apr 6. J Mol Med (Berl). 2018. PMID: 29626254
-
The human antiviral factor TRIM11 is under the regulation of HIV-1 Vpr.PLoS One. 2014 Aug 8;9(8):e104269. doi: 10.1371/journal.pone.0104269. eCollection 2014. PLoS One. 2014. PMID: 25105968 Free PMC article.
-
Inhibition of HIV early replication by the p53 and its downstream gene p21.Virol J. 2018 Mar 27;15(1):53. doi: 10.1186/s12985-018-0959-x. Virol J. 2018. PMID: 29587790 Free PMC article.
-
Degradation of SAMHD1 by Vpx Is Independent of Uncoating.J Virol. 2015 May;89(10):5701-13. doi: 10.1128/JVI.03575-14. Epub 2015 Mar 11. J Virol. 2015. PMID: 25762741 Free PMC article.
-
CRL4-DCAF1 Ubiquitin Ligase Dependent Functions of HIV Viral Protein R and Viral Protein X.Viruses. 2024 Aug 17;16(8):1313. doi: 10.3390/v16081313. Viruses. 2024. PMID: 39205287 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

