A conformation-dependent neutralizing monoclonal antibody specifically targeting receptor-binding domain in Middle East respiratory syndrome coronavirus spike protein
- PMID: 24719424
- PMCID: PMC4054355
- DOI: 10.1128/JVI.00433-14
A conformation-dependent neutralizing monoclonal antibody specifically targeting receptor-binding domain in Middle East respiratory syndrome coronavirus spike protein
Abstract
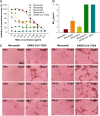
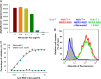
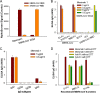
Prophylactic and therapeutic strategies are urgently needed to combat infections caused by the newly emerged Middle East respiratory syndrome coronavirus (MERS-CoV). Here, we have developed a neutralizing monoclonal antibody (MAb), designated Mersmab1, which potently blocks MERS-CoV entry into human cells. Biochemical assays reveal that Mersmab1 specifically binds to the receptor-binding domain (RBD) of the MERS-CoV spike protein and thereby competitively blocks the binding of the RBD to its cellular receptor, dipeptidyl peptidase 4 (DPP4). Furthermore, alanine scanning of the RBD has identified several residues at the DPP4-binding surface that serve as neutralizing epitopes for Mersmab1. These results suggest that if humanized, Mersmab1 could potentially function as a therapeutic antibody for treating and preventing MERS-CoV infections. Additionally, Mersmab1 may facilitate studies of the conformation and antigenicity of MERS-CoV RBD and thus will guide rational design of MERS-CoV subunit vaccines.
Importance: MERS-CoV is spreading in the human population and causing severe respiratory diseases with over 40% fatality. No vaccine is currently available to prevent MERS-CoV infections. Here, we have produced a neutralizing monoclonal antibody with the capacity to effectively block MERS-CoV entry into permissive human cells. If humanized, this antibody may be used as a prophylactic and therapeutic agent against MERS-CoV infections. Specifically, when given to a person (e.g., a patient's family member or a health care worker) either before or after exposure to MERS-CoV, the humanized antibody may prevent or inhibit MERS-CoV infection, thereby stopping the spread of MERS-CoV in humans. This antibody can also serve as a useful tool to guide the design of effective MERS-CoV vaccines.
Copyright © 2014, American Society for Microbiology. All Rights Reserved.
Figures


References
-
- Zhong NS, Zheng BJ, Li YM, Poon, Xie ZH, Chan KH, Li PH, Tan SY, Chang Q, Xie JP, Liu XQ, Xu J, Li DX, Yuen KY, Peiris Guan Y. 2003. Epidemiology and cause of severe acute respiratory syndrome (SARS) in Guangdong, People's Republic of China, in February, 2003. Lancet 362:1353–1358. 10.1016/S0140-6736(03)14630-2 - DOI - PMC - PubMed
-
- Mailles A, Blanckaert K, Chaud P, van der Werf S, Lina B, Caro V, Campese C, Guery B, Prouvost H, Lemaire X, Paty MC, Haeghebaert S, Antoine D, Ettahar N, Noel H, Behillil S, Hendricx S, Manuguerra JC, Enouf V, La RG, Semaille C, Coignard B, Levy-Bruhl D, Weber F, Saura C, Che D. 2013. First cases of Middle East respiratory syndrome coronavirus (MERS-CoV) infections in France, investigations and implications for the prevention of human-to-human transmission, France, May 2013. Euro Surveill. 18:pii=20502 http://www.eurosurveillance.org/ViewArticle.aspx?ArticleId=20502 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

