Conserved neutralizing epitope at globular head of hemagglutinin in H3N2 influenza viruses
- PMID: 24719430
- PMCID: PMC4054433
- DOI: 10.1128/JVI.00420-14
Conserved neutralizing epitope at globular head of hemagglutinin in H3N2 influenza viruses
Abstract
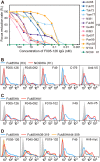
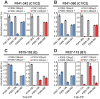
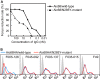
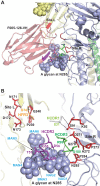
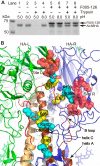
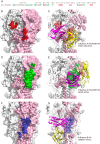
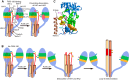
Neutralizing antibodies that target the hemagglutinin of influenza virus either inhibit binding of hemagglutinin to cellular receptors or prevent the low-pH-induced conformational change in hemagglutinin required for membrane fusion. In general, the former type of antibody binds to the globular head formed by HA1 and has narrow strain specificity, while the latter type binds to the stem mainly formed by HA2 and has broad strain specificity. In the present study, we analyzed the epitope and function of a broadly neutralizing human antibody against H3N2 viruses, F005-126. The crystal structure of F005-126 Fab in complex with hemagglutinin revealed that the antibody binds to the globular head, spans a cleft formed by two hemagglutinin monomers in a hemagglutinin trimer, and cross-links them. It recognizes two peptide portions (sites L and R) and a glycan linked to asparagine at residue 285 using three complementarity-determining regions and framework 3 in the heavy chain. Binding of the antibody to sites L (residues 171 to 173, 239, and 240) and R (residues 91, 92, 270 to 273, 284, and 285) is mediated mainly by van der Waals contacts with the main chains of the peptides in these sites and secondarily by hydrogen bonds with a few side chains of conserved sequences in HA1. Furthermore, the glycan recognized by F005-126 is conserved among H3N2 viruses. F005-126 has the ability to prevent low-pH-induced conformational changes in hemagglutinin. The newly identified conserved epitope, including the glycan, should be immunogenic in humans and may induce production of broadly neutralizing antibodies against H3 viruses.
Importance: Antibodies play an important role in protection against influenza virus, and hemagglutinin is the major target for virus neutralizing antibodies. It has long been believed that all effective neutralizing antibodies bind to the surrounding regions of the sialic acid-binding pocket and inhibit the binding of hemagglutinin to the cellular receptor. Since mutations are readily introduced into such epitopes, this type of antibody shows narrow strain specificity. Recently, however, broadly neutralizing antibodies have been isolated. Most of these bind either to conserved sites in the stem region or to the sialic acid-binding pocket itself. In the present study, we identified a new neutralizing epitope in the head region recognized by a broadly neutralizing human antibody against H3N2. This epitope may be useful for design of vaccines.
Copyright © 2014, American Society for Microbiology. All Rights Reserved.
Figures


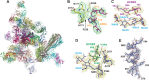



References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

