Integrative analysis of Salmonellosis in Israel reveals association of Salmonella enterica Serovar 9,12:l,v:- with extraintestinal infections, dissemination of endemic S. enterica Serovar Typhimurium DT104 biotypes, and severe underreporting of outbreaks
- PMID: 24719441
- PMCID: PMC4042803
- DOI: 10.1128/JCM.00399-14
Integrative analysis of Salmonellosis in Israel reveals association of Salmonella enterica Serovar 9,12:l,v:- with extraintestinal infections, dissemination of endemic S. enterica Serovar Typhimurium DT104 biotypes, and severe underreporting of outbreaks
Abstract
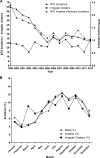
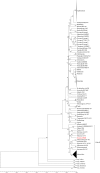
Salmonella enterica is the leading etiologic agent of bacterial food-borne outbreaks worldwide. This ubiquitous species contains more than 2,600 serovars that may differ in their host specificity, clinical manifestations, and epidemiology. To characterize salmonellosis epidemiology in Israel and to study the association of nontyphoidal Salmonella (NTS) serovars with invasive infections, 48,345 Salmonella cases reported and serotyped at the National Salmonella Reference Center between 1995 and 2012 were analyzed. A quasi-Poisson regression was used to identify irregular clusters of illness, and pulsed-field gel electrophoresis in conjunction with whole-genome sequencing was applied to molecularly characterize strains of interest. Three hundred twenty-nine human salmonellosis clusters were identified, representing an annual average of 23 (95% confidence interval [CI], 20 to 26) potential outbreaks. We show that the previously unsequenced S. enterica serovar 9,12:l,v:- belongs to the B clade of Salmonella enterica subspecies enterica, and we show its frequent association with extraintestinal infections, compared to other NTS serovars. Furthermore, we identified the dissemination of two prevalent Salmonella enterica serovar Typhimurium DT104 clones in Israel, which are genetically distinct from other global DT104 isolates. Accumulatively, these findings indicate a severe underreporting of Salmonella outbreaks in Israel and provide insights into the epidemiology and genomics of prevalent serovars, responsible for recurring illness.
Copyright © 2014, American Society for Microbiology. All Rights Reserved.
Figures

Similar articles
-
CRISPR-MVLST subtyping of Salmonella enterica subsp. enterica serovars Typhimurium and Heidelberg and application in identifying outbreak isolates.BMC Microbiol. 2013 Nov 12;13:254. doi: 10.1186/1471-2180-13-254. BMC Microbiol. 2013. PMID: 24219629 Free PMC article.
-
Presence of β-lactamases in extended-spectrum-cephalosporin-resistant Salmonella enterica of 30 different serovars in Germany 2005-11.J Antimicrob Chemother. 2013 Sep;68(9):1978-81. doi: 10.1093/jac/dkt163. Epub 2013 May 14. J Antimicrob Chemother. 2013. PMID: 23674765
-
Detection of multidrug-resistant Salmonella enterica serovar typhimurium phage types DT102, DT104, and U302 by multiplex PCR.J Clin Microbiol. 2006 Jul;44(7):2354-8. doi: 10.1128/JCM.00171-06. J Clin Microbiol. 2006. PMID: 16825349 Free PMC article.
-
Salmonella enterica Serovar Panama, an Understudied Serovar Responsible for Extraintestinal Salmonellosis Worldwide.Infect Immun. 2019 Aug 21;87(9):e00273-19. doi: 10.1128/IAI.00273-19. Print 2019 Sep. Infect Immun. 2019. PMID: 31262982 Free PMC article. Review.
-
Invasive Nontyphoidal Salmonella Infections Among Children in Mali, 2002-2014: Microbiological and Epidemiologic Features Guide Vaccine Development.Clin Infect Dis. 2015 Nov 1;61 Suppl 4(Suppl 4):S332-8. doi: 10.1093/cid/civ729. Clin Infect Dis. 2015. PMID: 26449949 Free PMC article. Review.
Cited by
-
Increasing incidence of invasive nontyphoidal Salmonella infections in Queensland, Australia, 2007-2016.PLoS Negl Trop Dis. 2019 Mar 18;13(3):e0007187. doi: 10.1371/journal.pntd.0007187. eCollection 2019 Mar. PLoS Negl Trop Dis. 2019. PMID: 30883544 Free PMC article.
-
Molecular docking-based virtual screening, drug-likeness, and pharmacokinetic profiling of some anti-Salmonella typhimurium cephalosporin derivatives.J Taibah Univ Med Sci. 2023 Jun 9;18(6):1417-1431. doi: 10.1016/j.jtumed.2023.05.021. eCollection 2023 Dec. J Taibah Univ Med Sci. 2023. PMID: 38162870 Free PMC article.
-
Comparative genomics of Salmonella enterica serovar Montevideo reveals lineage-specific gene differences that may influence ecological niche association.Microb Genom. 2018 Aug;4(8):e000202. doi: 10.1099/mgen.0.000202. Epub 2018 Jul 27. Microb Genom. 2018. PMID: 30052174 Free PMC article.
-
Complete genome sequence of multi-drug-resistant Salmonella enterica subsp. enterica serovar Typhimurium strain Bnaya isolated from a dairy calf in Israel.Microbiol Resour Announc. 2023 Oct 19;12(10):e0035123. doi: 10.1128/MRA.00351-23. Epub 2023 Oct 3. Microbiol Resour Announc. 2023. PMID: 37787537 Free PMC article.
-
From Eberthella typhi to Salmonella Typhi: The Fascinating Journey of the Virulence and Pathogenicity of Salmonella Typhi.ACS Omega. 2023 Jul 14;8(29):25674-25697. doi: 10.1021/acsomega.3c02386. eCollection 2023 Jul 25. ACS Omega. 2023. PMID: 37521659 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
Associated data
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

