Methylome diversification through changes in DNA methyltransferase sequence specificity
- PMID: 24722038
- PMCID: PMC3983042
- DOI: 10.1371/journal.pgen.1004272
Methylome diversification through changes in DNA methyltransferase sequence specificity
Abstract
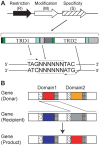
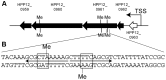
Epigenetic modifications such as DNA methylation have large effects on gene expression and genome maintenance. Helicobacter pylori, a human gastric pathogen, has a large number of DNA methyltransferase genes, with different strains having unique repertoires. Previous genome comparisons suggested that these methyltransferases often change DNA sequence specificity through domain movement--the movement between and within genes of coding sequences of target recognition domains. Using single-molecule real-time sequencing technology, which detects N6-methyladenines and N4-methylcytosines with single-base resolution, we studied methylated DNA sites throughout the H. pylori genome for several closely related strains. Overall, the methylome was highly variable among closely related strains. Hypermethylated regions were found, for example, in rpoB gene for RNA polymerase. We identified DNA sequence motifs for methylation and then assigned each of them to a specific homology group of the target recognition domains in the specificity-determining genes for Type I and other restriction-modification systems. These results supported proposed mechanisms for sequence-specificity changes in DNA methyltransferases. Knocking out one of the Type I specificity genes led to transcriptome changes, which suggested its role in gene expression. These results are consistent with the concept of evolution driven by DNA methylation, in which changes in the methylome lead to changes in the transcriptome and potentially to changes in phenotype, providing targets for natural or artificial selection.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures


References
-
- Poetsch AR, Plass C (2011) Transcriptional regulation by DNA methylation. Cancer Treat Rev 37 Suppl 1S8–12. - PubMed
-
- Suzuki MM, Bird A (2008) DNA methylation landscapes: provocative insights from epigenomics. Nat Rev Genet 9: 465–476. - PubMed
-
- Low DA, Casadesus J (2008) Clocks and switches: bacterial gene regulation by DNA adenine methylation. Curr Opin Microbiol 11: 106–112. - PubMed
-
- Oshima T, Wada C, Kawagoe Y, Ara T, Maeda M, et al. (2002) Genome-wide analysis of deoxyadenosine methyltransferase-mediated control of gene expression in Escherichia coli . Mol Microbiol 45: 673–695. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

