Discovery and validation of an INflammatory PROtein-driven GAstric cancer Signature (INPROGAS) using antibody microarray-based oncoproteomics
- PMID: 24722433
- PMCID: PMC4039123
- DOI: 10.18632/oncotarget.1879
Discovery and validation of an INflammatory PROtein-driven GAstric cancer Signature (INPROGAS) using antibody microarray-based oncoproteomics
Abstract
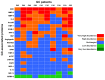
This study aimed to improve gastric cancer (GC) diagnosis by identifying and validating an INflammatory PROtein-driven GAstric cancer Signature (hereafter INPROGAS) using low-cost affinity proteomics. The detection of 120 cytokines, 43 angiogenic factors, 41 growth factors, 40 inflammatory factors and 10 metalloproteinases was performed using commercially available human antibody microarray-based arrays. We identified 21 inflammation-related proteins (INPROGAS) with significant differences in expression between GC tissues and normal gastric mucosa in a discovery cohort of matched pairs (n=10) of tumor/normal gastric tissues. Ingenuity pathway analysis confirmed the "inflammatory response", "cellular movement" and "immune cell trafficking" as the most overrepresented biofunctions within INPROGAS. Using an expanded independent validation cohort (n = 22), INPROGAS classified gastric samples as "GC" or "non-GC" with a sensitivity of 82% (95% CI 59-94) and a specificity of 73% (95% CI 49-89). The positive predictive value and negative predictive value in this validation cohort were 75% (95% CI 53-90) and 80% (95% CI 56-94), respectively. The positive predictive value and negative predictive value in this validation cohort were 75% (95% CI 53-90) and 80% (95% CI 56-94), respectively. Antibody microarray analyses of the GC-associated inflammatory proteome identified a 21-protein INPROGAS that accurately discriminated GC from noncancerous gastric mucosa.
Figures



References
-
- Jemal A, Siegel R, Ward E, Hao Y, Xu J, Murray T, Thun MJ. Cancer statistics, 2008. CA Cancer J Clin. 2008;58:71–96. - PubMed
-
- Mori M, Mimori K, Shiraishi T, Tanaka S, Ueo H, Sugimachi K, Akiyoshi T. p27 expression and gastric carcinoma. Nat Med. 1997;3:593. - PubMed
-
- Graziano F, Mandolesi A, Ruzzo A, Bearzi I, Testa E, Arduini F, Silva R, Muretto P, Mari D, Berardi R, Scartozzi M, Lai V, Cascinu S, Magnani M. Predictive and prognostic role of E-cadherin protein expression in patients with advanced gastric carcinomas treated with palliative chemotherapy. Tumour Biol. 2004;25:106–110. - PubMed
-
- Sanz-Ortega J, Steinburg SM, Moro E, Saez M, Lopez JA, Sierra E, Sanz-Esponera J, Merino MJ. Comparative study of tumor angio- genesis and immunohistochemistry for p53, c-erbB2, c-myc and EGFr as prognostic factors in gastric cancer. Histol Histopathol. 2000;15:455–462. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

