Low-dose formaldehyde delays DNA damage recognition and DNA excision repair in human cells
- PMID: 24722772
- PMCID: PMC3983121
- DOI: 10.1371/journal.pone.0094149
Low-dose formaldehyde delays DNA damage recognition and DNA excision repair in human cells
Abstract
Objective: Formaldehyde is still widely employed as a universal crosslinking agent, preservative and disinfectant, despite its proven carcinogenicity in occupationally exposed workers. Therefore, it is of paramount importance to understand the possible impact of low-dose formaldehyde exposures in the general population. Due to the concomitant occurrence of multiple indoor and outdoor toxicants, we tested how formaldehyde, at micromolar concentrations, interferes with general DNA damage recognition and excision processes that remove some of the most frequently inflicted DNA lesions.
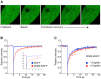
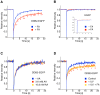
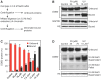
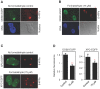
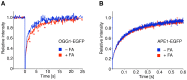
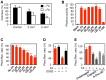
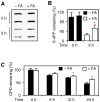
Methodology/principal findings: The overall mobility of the DNA damage sensors UV-DDB (ultraviolet-damaged DNA-binding) and XPC (xeroderma pigmentosum group C) was analyzed by assessing real-time protein dynamics in the nucleus of cultured human cells exposed to non-cytotoxic (<100 μM) formaldehyde concentrations. The DNA lesion-specific recruitment of these damage sensors was tested by monitoring their accumulation at local irradiation spots. DNA repair activity was determined in host-cell reactivation assays and, more directly, by measuring the excision of DNA lesions from chromosomes. Taken together, these assays demonstrated that formaldehyde obstructs the rapid nuclear trafficking of DNA damage sensors and, consequently, slows down their relocation to DNA damage sites thus delaying the excision repair of target lesions. A concentration-dependent effect relationship established a threshold concentration of as low as 25 micromolar for the inhibition of DNA excision repair.
Conclusions/significance: A main implication of the retarded repair activity is that low-dose formaldehyde may exert an adjuvant role in carcinogenesis by impeding the excision of multiple mutagenic base lesions. In view of this generally disruptive effect on DNA repair, we propose that formaldehyde exposures in the general population should be further decreased to help reducing cancer risks.
Conflict of interest statement
Figures

References
-
- Donaldson MR, Coldiron BM (2011) No end in sight: the skin cancer epidemic continues. Semin Cutan Med Surg 30: 3–5. - PubMed
-
- Hoeijmakers JH (2009) DNA damage, aging and cancer. New Engl J Med 361: 1475–1485. - PubMed
-
- Friedberg EC, Walker GC, Siede W, Wood RD, Schultz RA, et al. (2006) DNA Repair and Mutagenesis (ASM Press, Washington D.C).
-
- Parsons JL, Dianov GL (2013) Co-ordination of base excision repair and genome stability. DNA Rep 12: 326–333. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

